Real-time PCR detection of S. aureus in pharmaceutical products
Posted: 21 December 2018 | Arianna Pinto, Lisa Pincus, Luis Jimenez, Sibora Peca, Stephanie Perez | No comments yet
The contamination of pharmaceuticals by microorganisms represents a major threat to public health, not just in the USA and Europe, but all around the world. Pathogenic microorganisms cause morbidity and in some cases mortality when present in products.1,2 Spoilage due to microbial breakdown of formulations can affect potency and shelf life, yet non-sterile pharmaceuticals contain a microbial bioburden that is not detrimental to the formulation or hazardous to consumers.

Based upon compendial tests, when non-sterile pharmaceuticals are tested, objectionable microorganisms must be absent from the finished product and raw materials.1,2 For this reason, regulatory agencies around the world require the absence of certain types of microorganisms that can be considered hazardous to consumers or compromise drug stability and efficacy. Staphylococcus aureus is one of the indicator microorganisms that must be absent from non-sterile pharmaceutical drugs; however, based upon published scientific surveys, S. aureus is a frequent microbial contaminant in both sterile and non-sterile pharmaceutical products around the world.1-3 Furthermore, infections by S. aureus are the number one cause of nosocomial outbreaks in the USA,4 while common infections by S. aureus are related to skin eruptions, bacteremia, endocarditis, toxic shock syndrome and pneumonia.5 S. aureus is also characterised by the ability to develop resistance to almost any antibiotic.
The challenge for pharmaceutical manufacturers is to produce a non-sterile drug with low levels of microorganisms and a complete absence of the ‘wrong type’ of objectionable microbes.
Some guidance to determine what is objectionable is provided by the Code of Federal Regulations and the USP informational chapter <1111>.2 Most pharmaceutical companies have relied on traditional cultivation and phenotypic methods to isolate and identify microbial contaminants;1,6 however, these methods are time consuming (5–7 days) and laborious, and rely upon the growth of microorganisms on specific substrates in the media. Of note, some bacteria do not grow on those substrates or otherwise grow too slowly to be detected by the incubation times currently used. In some cases, microbial cells undergo a physiological state by reducing metabolic reaction rates and cell size, which affect the protein and enzymatic profiles used to identify environmental isolates.6 These microbial cell changes are triggered by physical processes and environmental systems implemented to reduce or eliminate microorganisms during pharmaceutical manufacturing. If the processes and systems are not validated or properly implemented, microorganisms can contaminate products, raw materials and equipment.1,2
Conventional methods relying on selective agar media after enrichment in tryptic soy broth (TSB) containing 2 percent Tween 20 were used to detect S. aureus in pharmaceutical samples with low levels of bacterial contamination – eg, below 75 colony-forming units (CFU). The selected bacteria were commonly found in products recalled in the USA.1,2 Following inoculation, samples were shaken for 18–24 hours at 37°C to promote bacterial growth. Table 1 shows the results for each selective agar media analysed: phenylethyl alcohol (PEA) and mannitol salt agar (MSA). All seven products tested were found to show bacterial growth. Identification of colonies growing on selective agar media for Gram-positive bacteria, PEA and MSA, showed S. aureus. However, the antiseptic mouthwash did not show any bacterial growth on PEA but S. aureus was isolated and identified on MSA.
Bacterial DNA was then extracted from all contaminated products. To determine the predominant bacterial DNA in the extracted product suspensions, a regular PCR reaction was performed targeting the 1.5 kilobases (kb) 16S rRNA eubacterial gene – a universal gene present in all members of the domain bacteria.7 The gene has conserved and variable regions that can be used to distinguish bacterial phyla, genera and species. The amplified fragments from product suspensions were cloned into vector plasmid pCR®4-TOPO. DNA sequencing of the clones demonstrated the predominant presence of E. coli in the clone library, but no other bacterial 16S rRNA sequence was detected.
To specifically detect S. aureus in the extracted bacterial DNA, a 273bp fragment of the S. aureus 16S rRNA gene was used as a target to develop the real-time PCR assay.8 Real-time PCR was performed with a LightCycler System9 using SYBR green PCR master mix as the detection system in a reaction mixture of 25ul. The copy number for S. aureus 16S rRNA gene is either five or six.10 The DNA concentration for undiluted samples of S. aureus DNA was found to have 1,144 nanograms (ng) per ml with a Cq of 10.13 (Table 2). The Cq indicated the cycle at which fluorescence from amplification exceeded the background fluorescence. When 10-fold dilutions of S. aureus DNA were tested, the minimum concentration detected was 0.01144ng/ml with a correlation coefficient of 0.98 (Figure 1). The Cq for the lowest DNA concentration detected was 27.04. Melting curve analysis of the amplified fragments confirmed the presence of the 273bp fragment.
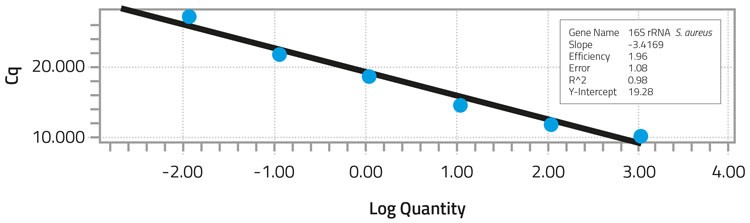

Standard curve analysis of S. aureus DNA dilutions after real-time PCR
All contaminated products analysed were found to have Cq values ranging from 16.80 to 26.29. The faster Cq values were found with the Hydrocodone Acetaminophen tablets, while Acetaminophen tablets exhibited for the longest amount of time. This indicated that the growth conditions and neutralisation protocol provided optimal conditions for the low numbers of S. aureus to grow sufficiently in the presence of other bacteria. S. aureus DNA can therefore be extracted from the bacterial mixed culture using a mild DNA extraction protocol and subsequently detected by real-time PCR. The low S. aureus inoculum of 24CFU/ml was easily detected after a 24-hour enrichment with shaking at 37°C.
Conclusions
A real-time PCR assay was developed to detect S. aureus in pharmaceutical products contaminated with low numbers of bacterial contamination by Burkholderia cepacia, Escherichia coli, S. aureus, and Bacillus megaterium. All samples showing positive results with standard microbiological methods were also positive with real-time PCR. Rapid assessment of pharmaceutical samples provides important information for quality control and highlights the possible risk of contaminated material, allowing the expeditious implementation of corrective actions to prevent morbidity and mortality by the lack of process control during manufacturing of pharmaceutical products.
References
1. Jimenez L. Microbial diversity in pharmaceutical product recalls and environments. PDA Journal of Pharmaceutical Science and Technology. 2007; 61:383-399.
2. Sutton SW, Jimenez L. A review of reported recalls involving microbiological control 2004- 2011 with emphasis on FDA considerations of “objectionable organisms”. American Pharmaceutical Review. 2012 Jan/Feb; 15:42-57.
3. Essam Eissa M. Distribution of bacterial contamination in non-sterile pharmaceutical materials and assessment of its risk to the health of the final consumers quantitatively. Beni-Suef University Journal of Basic and Applied Sciences. 2016; 5:217-230.
4. Bischoff WE, Wallis ML, Tucker KB, Reboussin BA, Sherertz RJ. Staphylococcus aureus nasal carriage in a student community, prevalence, clonal relationships, and risk factors. Infection Control and Hospital Epidemics. 2004; 25:485-491.
5. Jimenez L, Kulko E, Barron E. Molecular characterization and antimicrobial susceptibility of Staphylococcus aureus isolates from a healthy student population. J. Microbiol Exp. 2014; 1(5): 00030. doi:10.15406/jmen.2014.01.00030.
6. Jimenez L. Molecular applications to pharmaceutical processes and clean room environments. PDA Journal of Pharmaceutical Science and Technology. 2011; 65:242-253.
7. Vetrovsky T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE. 2013; 8(2): e57923. doi:10.1371/ journal.pone.0057923.
8. Saruta K, Hoshina S, Machida K. Genetic identification of Staphylococcus aureus by polymerase chain reaction using single-base-pair mismatch in 16S ribosomal RNA gene. Microbiol. Immunol. 1995; 39:839-844.
9. Jimenez L, Jashari T, Vasquez J, Zapata S, Bochis J, Kulko M, Ellman V, Gardner M, Choe T. Real-Time PCR detection of Burkholderia cepacia in pharmaceutical products contaminated with low levels of bacterial contamination. PDA Journal of Pharmaceutical Science and Technology. 2018; 72:73-80.
10. Fluit AC, Jansen MD, Bosch T, Jansen WTM, Schouls L, Jonker MJ, Boel CHE. rRNA operon copy number can explain the distinct epidemiology of hospital-associated methicillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemoteraphy. 2016; 60:7313-7320.
Biography










The rest of this content is restricted - login or subscribe free to access


Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- bi-monthly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Issue
Related topics
Analytical techniques, Drug Development, Drug Manufacturing, Drug Safety, Formulation, Microbial Biological Manufacturing, Microbial Detection, Microbiology, Polymerase Chain Reaction (PCR)
Related organisations
Related people
Arianna Pinto, Lisa Pincus, Luis Jimenez, Sibora Peca, Stephanie Perez