Subvisible particle identification in protein-based formulations by Raman spectroscopy
Posted: 29 June 2017 | Markus Lankers (Rap.ID), Olga Laskina (Rap.ID), Oliver Valet (Rap.ID) | No comments yet
Subvisible particles in protein-based formulations can have different origins. Particles can be extrinsic (unexpected foreign material), intrinsic (from the production environment or primary packaging), and inherent (from the formulation). It is important that inherent particles (protein agglomerates) are distinguished from the other two types and that the extrinsic and intrinsic particles sources are found and eventually eliminated…


Micro-flow imaging (MFI) technique allows quantification and characterisation of sub-visible particles based on the morphology, but lacks chemical identification.^1 Comparison of membrane microscopy and MFI methods indicates the underestimation of the number of the silicone droplets by counting on a membrane. Sample preparation of amorphous protein particles on a filter is challenging as it often leads to protein passing through the filter. Often, protein sample is stressed to cause coagulation and agglomeration prior to the deposition on a filter. However, this can alter protein structure and properties and is not a suitable method when characterisation of the pristine protein is desired. Therefore there is a need for in situ techniques for analysis of proteins.
Here the use of a sample cell for particles immersed in liquids, called ‘wet cell’, will be described. The wet cell allows the analysis of samples in situ. The sample in the wet cell is evaluated with a Raman microscope (Single Particle Explorer SPE) to determine size, location and further morphological details of micrometer particles in the suspension and performs their further Raman analysis. The use of Raman spectroscopy and a wet cell enables characterisation of particles in situ and helps to avoid sample preparation that can introduce undesired changes to the samples. This gives a direct link between shape information and chemical composition. Raman spectroscopy is the preferred tool to investigate particles in aqueous solutions as the water does not interfere with the spectroscopic information of the particles due to its weak vibrational features. The wet cell concept connects MFI shape information with chemical characterisation through Raman spectroscopy. This can generate valuable information on the pathways of particle formation, as well as stability data.
The idea of investigating particles in the formulation without filtration, and therefore keeping the characteristics of the particles is straightforward. But there are a number of experimental hurdles connected with the practical implementation. The different interfaces between the laser beam and the particles give a certain contribution to the resulting Raman spectra. If one considers a protein solution in a liquid cell made out of glass one can observe contributions from water, glass and protein solution. The use of a confocal setup can overcome this issue to some extent. The observation of protein particles in a concentrated protein solution usually requires a very good signal-to-noise ratio, as the spectral differences are quite small. The movement of particles in the cell, even in the micrometer scale, has to be avoided to get only the particle signal. This can be done using a very thin liquid layer of 20-200µm, and subsequently using a large area enabling a sufficient sampling volume. Figure 1 shows the wet cell setup for the Raman spectroscopic investigation. The liquid layer is 100µm thin and has a diameter of 40mm.
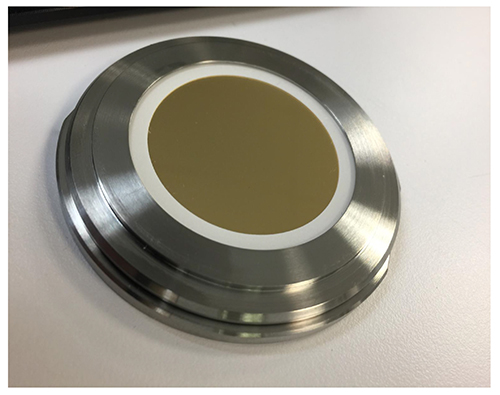

Figure 1: Wet cell setup for the Raman spectroscopic investigation.
Figure 2 shows the comparison of an image of protein particle captured in the wet cell and a comparable protein particle imaged with a Flowcam. The yellow background colour is due to the gold coating of the wet cell. Both images are of a comparable quality. At the same time, a chemical identification is possible in the wet cell by Raman spectroscopy.
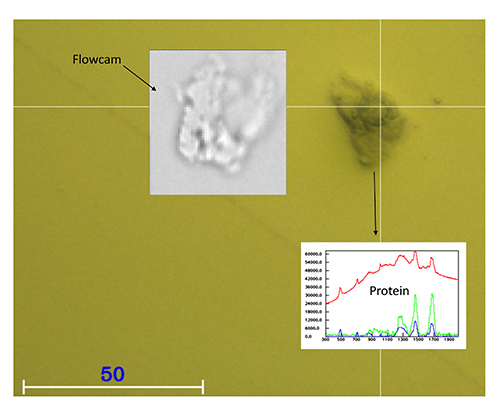

Figure 2: Comparison of an image of a protein particle captured in the wet cell and a comparable protein particle imaged with a flowcam.
The degradation of polysorbate and silicone oil aggregation are chosen as examples. Polysorbate is a surfactant commonly used in protein pharmaceuticals to stabilise formulations. Degradation of polysorbate causes the formation of fatty acids and results in a decrease in polysorbate. This leads to potential problems with protein stability due to decreased protection against stress.2
Silicone-induced protein aggregation is another common pathway of inherent particle formation. Silicone oil is often applied to the inner surface of syringes to form lubricating films. However, protein-based therapeutics can have strong interactions with the silicone oils. Excessive silicone or its heterogeneous distribution in syringes can induce protein aggregation in formulations.3-5
Two formulations were used in this study: a) monoclonal antibody stored beyond its shelf life; b) monoclonal antibody stored beyond its shelf life in a syringe. 600µL of each sample was placed into a wet cell, and then the cell was placed on the motorised stage of the Raman microscope. An area of a sample was scanned in an automated regime, images of the particles were taken with size and morphology analysis, and then particles were analysed with Raman spectroscopy at 532 nm excitation with 30s integration time to obtain spectra that allow material identification.
The results are summarised in Figure 3. Sample A indicates the presence of protein aggregates along with small fatty acid particles formed upon polysorbate degradation. Protein particle formation in sample A was caused by polysorbate degradation. Sample B shows protein particles in addition to multiple circular droplets that were identified by Raman spectroscopy as silicone oil. Protein aggregation in sample B was caused by silicone oil interaction with the protein-based formulation.
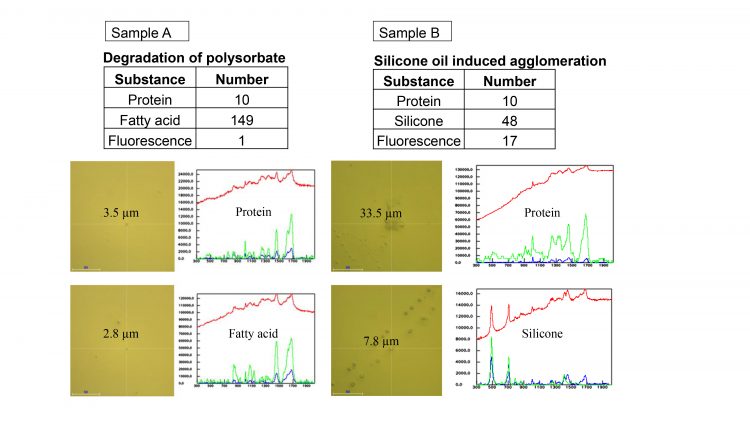

Figure 3: Results summarised. The table shows the number of particles identified in each of the samples.
The use of the wet cell enables characterisation of pristine protein particles and helps to avoid sample preparation that can introduce undesired changes to the samples. This helps to correlate shape, morphological information, and chemical composition acquired through Raman spectroscopy.
Biographies






References
- Strehl, R, Rombach-Riegraf V, Diez M, Egodage K, Blueme, M, Jeschke M, Koulov A. Discrimination between silicone oil droplets and protein aggregates in biopharmaceuticals: a novel multiparametric image filter for sub-visible particles in microflow imaging analysis. Pharmaceutical Research. 2012;29(2):594-602.
- Siska C, Pierini C, Lau H, Latypov R, Fesinmeyer R, Litowski J. Free Fatty Acid Particles in protein formulations, part 2: contribution of polysorbate raw material. Journal of Pharmaceutical Science. 2015;104(2):447-456.
- Li J, Pinnamaneni S, Quan Y, Jaiswal A, Andersson F, Zhang X. Mechanistic understanding of protein-silicone oil interactions. Pharmaceutical Research. 2012; 29(6):1689-1697.
- Gerhardt A, Mcgraw N, Schwartz D, Bee J, Carpenter J, Randolph T. Protein aggregation and particle formation in prefilled glass syringes. Journal of Pharmaceutical Science. 2014;103(6);1601-1612.
- Jones, L, Kaufmann A, Middaugh C. Silicone oil induced aggregation of proteins. Journal of Pharmaceutical Science. 2005;94(4);918-927.









