Digital polymerase chain reaction; new diagnostic opportunities
Posted: 22 February 2010 | Dr Jim Huggett, Molecular and Cell Biology, LGC and Dr Daniel J Scott, Project Manager, Research and Technology Division, LGC | No comments yet
LGC is an international science-based company located in South West London. A progressive and innovative enterprise, LGC operates in socially responsible fields underpinning the health, safety and security of the public, and regulation and enforcement for UK government departments and blue chip clients. Our products and services enable our customers to have a sound basis on which to base their scientific and commercial decisions or conformity to international statutory and regulatory standards.
LGC is an international science-based company located in South West London. A progressive and innovative enterprise, LGC operates in socially responsible fields underpinning the health, safety and security of the public, and regulation and enforcement for UK government departments and blue chip clients. Our products and services enable our customers to have a sound basis on which to base their scientific and commercial decisions or conformity to international statutory and regulatory standards.
LGC is an international science-based company located in South West London. A progressive and innovative enterprise, LGC operates in socially responsible fields underpinning the health, safety and security of the public, and regulation and enforcement for UK government departments and blue chip clients. Our products and services enable our customers to have a sound basis on which to base their scientific and commercial decisions or conformity to international statutory and regulatory standards.
Innovation, which is key to our continued success, underpins our lead role as the designated UK’s National Measurement Institute (NMI) for chemical and bio analytical measurement and UK’s Government Chemist (GC) function. It further drives both strategic and tactical research and development across the organisation. This role means that LGC is perfectly positioned at the forefront of new technological application, where a unique approach to the polymerase chain reaction (PCR) that has the potential to considerably advance molecular measurement may be about to become established.
Historically, there are many examples where ideas have preceded the technology needed to eventually enable their realisation. The use of PCR as a quantitative tool was being explored in the early 90s, but when compared with todays practice this was complex and inaccurate with a limited dynamic range1. It was not until earlier this decade that the use of fluorescent reporters and real time PCR instruments became more widely available enabling real time PCR to become fully established. Today real time PCR instruments are as common a laboratory instrument as conventional thermal cyclers were 10 years ago and molecular quantification is one of principle uses of the PCR technique.
In 1999 Vogelstein and Kinzler reported a concept to improve the sensitivity of PCR to enable detection of rare minority mutant sequences2. They were investigating rare SNP mutations in stool samples from colon cancer patients. Traditional legacy PCR was not sensitive enough for this application, due to the predominance of signal from the normal unmutated sequence. They devised a method that would increase the probability of detecting the rare mutant sequence using limiting dilution to partition the sample and isolate single templates into individual PCR reactions. When a PCR product was detected it was either mutated or normal. Subsequently, a simple sum of all reactions that produced a PCR product gave a direct read-out of the number of copies present in the original sample.
This approach transformed the linear signal of PCR into a digital, or binary, ‘yes’ or ‘no’ read-out (see Figure 1). Ironically this also required one of the most complex and tricky protocol setups yet devised for molecular biology. Since then this potentially brilliant idea has been explored in a number of specialist scenarios mostly in cancer research3, but has essentially been waiting in the wings for a technological advancement that would make this approach practically more amenable. The recent development in microfluidics is the technological advancement that will make digital PCR (dPCR) increasingly more amenable, possibly having a similar impact on real time PCR that the latter had on PCR. Microfluidic dPCR functions by distributing one reaction into hundreds of individual reactions chambers which reside on a microfluidic chip (see Figure 2A). This automated method also employs miniaturisation of the PCR so that hundreds of individual reactions are performed and analysed (see Figure 2B) from what is effectively a single PCR reaction.
dPCR has a number of application improvements over real time PCR, but there are three key benefits:
1. Absolute molecular counting!
dPCR’s strength is that it can provide “real” absolute quantity measurements. Real time PCR, using either relative or “absolute” methodologies, is actually a relative quantification approach, being compared relative to another sample or external standard. Thus the actual amount being measured is either not considered (which is statistically undesirable) or is compared to a calibrator of “known” amount, calculated using other means.
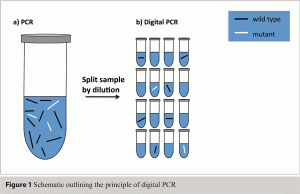

Figure 1 Schematic outlining the principle of digital PCR
One of the most established diagnostic applications of real time PCR is found in the field of virology where pathogens with small, frequently stable, genomes lend themselves to existing approaches. Due to the relative nature of real time PCR, laboratories compare results using centrally derived, globally distributed international units (IU), which are included alongside each analysis. While this works well, introduction of tests for newer viruses, or variants of existing ones are hindered by the subsequent need for the development and distribution of a new appropriate IU. Such an approach is also less suited to diagnosis of pathogens with larger genomes or quantification of patient nucleic acids in the advancing field of biomarker analysis.
As dPCR is actually counting the number of molecules present within a sample it can potentially be performed in different laboratories and directly compared. Thus, copies per ml of blood (for example) could be reported independent of the need for an IU to assign the result a value for comparison. While controls would be necessary for internal and external quality assurance, as with any diagnostic test, the absence of the need for an IU has the potential to vastly simplify the comparison of data. This would concomitantly increase the potential for rapidly translating novel diagnostic tests for different diseases, speeding up the subsequent patient impact.
2. Measurement like never before
dPCR also opens a window of detection sensitivity that is unprecedented, potentially detecting minute changes in gene sequence or quantity. At LGC we have pushed dPCR to detect differences of less than 1.3 fold. Such precision has the potential for measuring somatic aneuploidy where the mutant sequence forms less than 10 % of the total somatic DNA. This method opens the possibility of non invasive tumour identification (e.g. through the analysis of blood, urine, sputum or stool samples) at a much earlier disease stage with possible improved diagnosis leading to better prognosis than is currently possible.
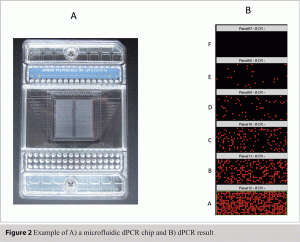

Figure 2 Example of A) a microfluidic dPCR chip and B) dPCR result
Another potential benefit of this approach is prenatal screening for foetal chromosomal abnormalities in maternal blood. Foetal DNA represents up to 20% of maternal circulating nucleic acids in the plasma or serum (CNAPS)4. Using such an approach has no risk when compared with conventional invasive methods like Chorionic villus sampling or amniocentesis, which convey a risk of foetal miscarriage. The accuracy of dPCR can also theoretically be used for foetal genotyping using maternal blood as deviations of expected maternal zygosity are attributed to the foetus and can be used to predict foetal genotype. This could provide considerable clinical value for assessing risk of single gene disorders such as cystic fibrosis or muscular dystrophy. Non-invasive detection of chromosomal aneuploidies such as Down’s syndrome (i.e. 3 copies of chromosome 21) is beyond current dPCR methodologies5. However, this could well be overcome in the near future with improved extraction methods capable of foetal DNA enrichment from blood samples and/or increased levels of replication during sample partitioning which can reliably differentiate even more subtle changes in gene/chromosome copy number.
3. Trace analysis
dPCR enables the detection of trace level nucleic acid targets. This is because sample partitioning not only isolates individual templates, but also increases their signal to noise ratio, effectively enriching DNA targets that were present at very low levels in the original sample. It was for this reason, as discussed above, that dPCR was initially developed2. This application has potential impact for numerous scenarios including tumour mutation detection, tumour or pathogen resistance screening as well as super infection analysis. dPCR could also potentially improve foetal mutation analysis and sexing, again when using maternal CNAPS, and when combined with reverse transcription provide a range of unique opportunities for cDNA analysis. Another advantage of sample partitioning is that the process of dilution also reduces sample matrix effects, like inhibition, that can alter the result6.
Last thoughts
The development of microfluidics that facilitate dPCR, while increasing the number of reactions actually conducted, will also greatly reduce reagent costs. It is possible, with current technology, to perform 4-8 µl PCR reactions which are divided into hundreds of minute reactions. This required volume is likely to reduce with time and the cost of the hardware, which is currently high, will also decrease; today’s qPCR machines are one tenth of the cost they were when they first appeared in the UK over a decade ago.
Finally the microfluidics setup also goes hand in hand with the automation of data acquisition, which is already one of the major benefits of existing real time PCR. However, real time PCR has suffered from ambiguity frequently associated with the subsequent analysis of data8. This is mainly due to the fact that the quantification cycle or Cq (formerly Ct or Cp) is a completely arbitrary measurement. Unlike real time PCR, dPCR better lends itself to automation of the subsequent analysis, due to the digital nature of the procedure which is independent of Cq. As with all PCR, dPCR will require optimisation for best results; this is something that has not yet been discussed widely. At LGC, we have an on-going programme of work funded by the National Measurement Office (as part of the UK National Measurement System Chemical and Biological Metrology Programme) to evaluate the technical performance of dPCR and, importantly, to understand the key factors that may influence assay performance and cause measurement bias. Our findings will be rapidly disseminated to ensure that this valuable technique is able to maximise its potential as a rapid and accurate diagnostic tool.
References
- Ferre F. Quantitative or semi-quantitative PCR: reality versus myth. PCR Methods Appl. 1992 Aug;2(1):1-9
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):9236-41.
- Pohl G, Shih IeM. Principle and applications of digital PCR. Expert Rev Mol Diagn. 2004 Jan;4(1):41-7
- Lun FM, Chiu RW, Allen Chan KC, Yeung Leung T, Kin Lau T, Dennis Lo YM. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008 Oct;54(10):1664-72
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008 Oct 21;105(42):16266-71.
- Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, Zumla A. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes. 2008 Aug 28;1:70
- Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006 Jul;4(7):e204.
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe, et al. (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 8(12):E63.
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009 Apr;55(4):611-22.




