Applications in bioprocesses and biotechnology
Posted: 11 November 2005 | Roger M. Jarvis (University of Manchester), Royston Goodacre (University of Manchester) | No comments yet
Raman spectroscopy is a highly versatile tool that provides chemical fingerprints from biological material that can be interpreted using chemometrics and machine learning. In combination this powerful approach is being developed for the quantitative determination of multiple determinands in bioprocesses and for the characterisation of microorganisms.
Many of today’s drugs arise from bioprocesses including microbial, yeast and animal cell lines. These include small molecules such as antibiotics and the so-called biopharmaceutical (recombinant) proteins and gene therapeutics. Indeed, currently the biopharmaceutical market is estimated to grow by 20% per year compared with 7 to 8% for the overall drug market.
The ability to control a bioprocess is paramount for product yield optimisation and it is imperative that the concentration of the fermentation product, the determinand, is assessed accurately. Whilst the development of such monitoring methods was traditionally driven by economic and ecological needs, the increasing manufacture of biopharmaceuticals has raised concerns over product consistency. For example, the glycosylation status of a protein must be known and protein purity is also highly important.
Thus, within the pharmaceutical sector there is an increasing need to embrace PAT and – particularly with the FDA poised to make this mandatory – Europe will inevitably follow. This should not, of course, be the sole reason for implementing PAT. PAT will allow for improvements in the biological understanding of a bioprocess with respect to cellular processes. This could then be used for bioprocess design and metabolic engineering to yield enhanced bioprocessing.
Within medicine there is a continuing – and in some areas urgent – need to find new APIs. Combinatorial chemistry has not produced any blockbusters and so this needs to be improved. There is still a trend to replace traditional chemical processes with bioprocesses because of their chemical specificity, including chirality, desirable reaction kinetics and benign conditions. Since many drugs originate from natural products, or derivatives, there is still a need for rapid and efficient methods for the screening of large numbers of, for example, microbial cultures for the production of biologically active metabolites. During the last 20 years the rate at which new antimicrobial agents are produced has decreased dramatically, with alarming increases in the number of pathogens that are becoming multidrug resistant. Together these have created a patient healthcare risk and this is of great concern. A crucial aspect for the discovery of new antibiotics is the development of new techniques that allow rapid and accurate characterisation of the mode of action of the pharmacophore.
Raman spectroscopy
As an analytical tool Raman spectroscopy has many advantages against infrared spectroscopy, as well as other techniques, which make it attractive for the analysis of bioprocesses. The Raman measurements are non-destructive, non-intrusive and require little or no sample preparation. The low levels of interference that arise from water allow readings to be taken of analytes in aqueous solution with minimum interference. In addition, with the confocal nature of Raman spectroscopy, there is no need for the introduction of a probe into a fermentor as measurements can be made through windows in the side of the reaction vessel. Coupled with fibre optics, which in the visible can be 100m long, this makes Raman spectroscopy a very versatile and powerful tool for monitoring bioprocesses.
Raman microscopy measures the exchange of energy with electromagnetic radiation of a particular wavelength generated from a laser (Figure 1). This exchange of energy results in a measurable Raman shift in the wavelength of the incident laser light (or what is also known as the inelastic light scattering effect). Measurements are usually collected in a few minutes resulting in the construction of a Raman ‘fingerprint’ of the same sample under interrogation.
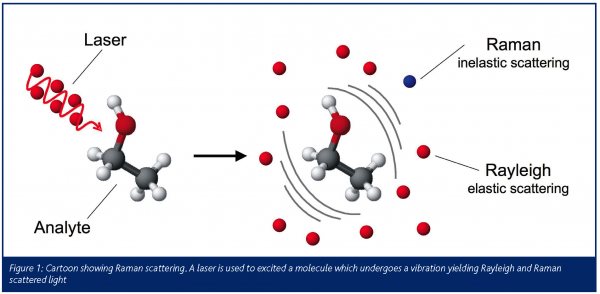

However, despite the apparent versatility of Raman spectroscopy, the Raman effect is particularly weak and typically only ~ 1 in 106-108 incident photons are inelastically scattered, thus collection times are in the order of minutes rather than seconds even when high laser power is used. Fortunately, the inelastic light scattering process can be enhanced (see Table 1) by using either resonance Raman (RR) spectroscopy or surface enhanced Raman spectroscopy (SERS) – methods that we have been developing for the analysis of bioprocesses and for the characterisation of microorganisms.
Chemometrics and machine learning
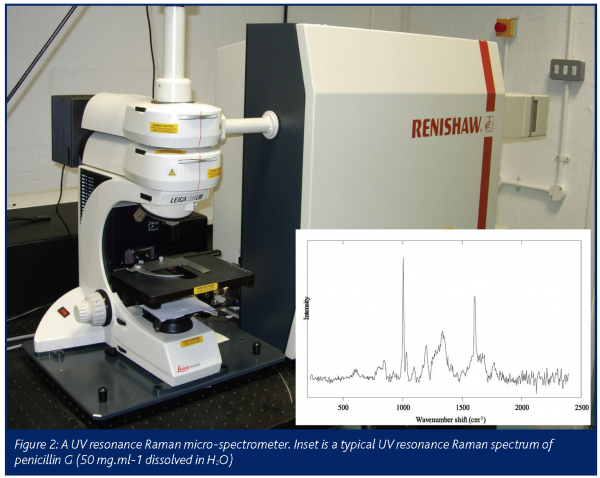
Raman spectra are very complex and contain information on all molecules in a biological sample. Figure 2 shows the UVRR spectrum of a relatively simple molecule which contains many vibrational modes. It is therefore impossible to use simple ‘stare and compare’ approaches and alternative strategies are required.

For the multivariate analysis of Raman spectra there are many tools in the chemometrics ‘zoo’. These approaches can loosely be split into the following three levels:
i. The clustering variety – These algorithms are based on unsupervised learning and seek to answer the question “How similar to one another are these samples based on the Raman fingerprints I have collected?” Typically principal components (PCA) and hierarchical cluster analyses (HCA) are used where the main goal is to check for reproducibility across the whole Raman spectra. In post-genomics such methods are sometimes referred to as ‘guilt-by-association’ where samples of known origin co-cluster with those of unknown provenance.
ii. The supervised learning variety – These algorithms are particularly pertinent for the analysis of bioprocesses and are based on supervised learning. These algorithms (vide infra) are calibrated (taught) and seek to give answers of biological interest that have much-lower dimensionality, such as ‘what are the levels of these metabolites in my bioprocess?’. For Raman spectroscopy partial least squares (PLS) regression is most popular. In addition, these can be used for classification where one wants to predict the class membership of something held in a Raman database of bacteria, for example. Typically discriminant function analysis (DFA) or the machine learning methods of artificial neural networks (ANNs) are used for this process.
iii. The inductive/mining variety – These machine learning algorithms are also based on supervised learning but, in addition to the above, seek to answer the question ‘What have I measured in my Raman fingerprint that makes samples in class A different from samples in class B?’ These variable selection algorithms are typically based on evolutionary computational methods of genetic algorithms (GAs) and genetic programming (GP), as well as classification and regression trees (CART).
Armed with the correct Raman spectroscopy and the appropriate chemometrics approach one can start to analyse bioprocesses and characterise microorganisms. Below are some examples of the Raman-chemometrics approaches we have been exploring with biotechnology.
Quantitative analysis of bioprocesses
Raman spectroscopy has historically been used to monitor the reaction of chemical processes and it was only a matter of time before this approach was used to analyse bioprocesses. Whilst chemical reactions involve few chemical species and vibrational modes more readily attributable to substrates and products, biological processes are more complex and are rarely vibrations directly related to bioproduct formation. Therefore for the quantitative analysis of (multiple) determinands in bioprocesses, PLS is used to correlate the ‘holistic’ Raman fingerprint with the level of a particular substrate or product. As this involves multivariate calibration the primary reference data (‘gold standard’ data typically involving HPLC(-MS)) to which the Raman signal will be correlated must be highly accurate. This means that the predictions from the Raman analyses will only be as good as the gold standard data. Despite this possible limitation, and as described above, due to its confocal nature it is possible to focus the laser into a liquid sample through a window in the fermentation vessel, rather than introducing a probe (as is normally required for near infrared), and generate data in real time so that the progression of the bioprocess can be monitored.
Our own studies have demonstrated that Raman spectroscopy can indeed be used for the on-line determination of the biotransformation by yeast of glucose to ethanol (Shaw et al., 1999). In this study both the concentrations of the substrate and product were accurately assessed and this allowed the progression of this microbial fermentation to be performed. Subsequently work also compared Raman spectroscopy with FT-IR and pyrolysis-MS for the quantification of gibberellic acid. In this application, a diverse range of unprocessed, industrial fed-batch fermentation broths containing the fungus Gibberella fujikuroi producing the natural product gibberellic acid, were directly analysed. It was concluded that Raman spectroscopy would seem to be the ideal bioprocess monitoring method as it is rapid, on-line, non-intrusive and gives interpretable answers of good precision (McGovern et al., 2002). More recently we demonstrated that SERS, using silver colloids, could be used for quantifying low levels of secondary metabolites in microbial processes (Clarke et al., 2005). Clearly this would be performed off-line due to the poisonous nature of the silver colloid on the microbial process, it would also be very unwise to add the colloid into the process plant!
Characterisation of microorganisms
For routine purposes the ideal method for microbial characterisation would have minimum sample preparation; analyse samples directly; be rapid, automated and (at least relatively) inexpensive. Clearly Raman spectroscopy has a role to play here and has indeed been explored (for a review see Maquelin et al., 2002). However, owing to the weak Raman effect several enhancement methods have been explored (Table 1).


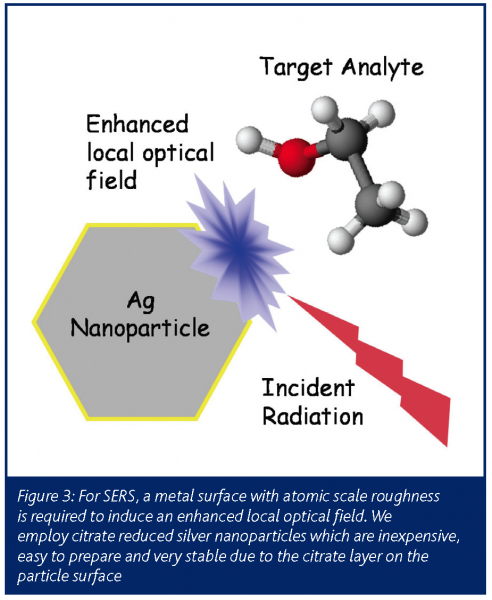
We have pioneered the rapid discrimination of bacteria using SERS and have recently shown, for the first time (Jarvis and Goodacre, 2004), that this approach is reproducible and has sufficient resolving power to discriminate between different strains of the same species and show with chemometrics that sub-species identification is possible. A cartoon of the SERS process is shown in Figure 3. The enhancement works on the nanometer range and, as a result, it is very likely that this method probes the surface of the bacteria and indeed our results suggest this to be the case. Moreover, owing to this and the very strong enhancements we have demonstrated that it is possible to collect information-rich SERS spectra from single bacterial cells in only a few seconds.

In addition to the rapid identification of bacteria we have very recently shown that it is also possible to use Raman spectroscopy to monitor the mode of action of antibiotics (López-Díez et al., 2005). In this study we investigated the sub-inhibitory effects of amikacin on Pseudomonas aeruginosa using UVRR. UVRR spectra were able to predict the concentration of amikacin to which bacterial cells had been exposed and 2D correlation spectroscopy contours maps indicated that spectral changes, due to the presence of amikacin in the growth media, occur according to the known mode of action of this antibiotic. We believe that this would be a powerful approach for the development and screening of new antibiotics, hopefully with novel modes of action.
Outlook
Raman spectroscopy clearly presents itself as a highly versatile tool that provides chemical fingerprints from biological material. Feasibility studies have demonstrated the ability to correlate Raman fingerprint with substrates and products in a similar fashion to near infrared spectroscopy. However, Raman has several advantages in that fibre optics can be readily coupled to the confocal nature of interrogation so that a probe-free non-intrusive analysis is performed. In addition, the Raman spectra are sharp as the fundamental vibration is seen rather than some broad overtone or harmonic as in near infrared. With the move towards full PAT on bioprocesses and biopharmaceuticals Raman will clearly have a role for the in-line monitoring and optimisation of the fermentations.
Raman spectroscopy also has the exquisite sensitivity required for the identification of microorganisms and there are already reports of single cell analysis. With the production of robust databases this would be highly attractive for the immediate assessment of bioprocess contamination. However, what is perhaps more exciting is the rapid identification of bacteria without prior cultivation, which would have implications in primary health care. Finally, the ability to correlate Raman spectra with the mode of action of pharmacophores will have use for the screening of novel bioactive compounds.
Acknowledgements
We would like to thank the BBSRC for funding.
References
Clarke, S.J., Littleford, R.E., Smith, W.E. & Goodacre, R. (2005) Rapid monitoring of antibiotics using Raman and surface enhanced Raman spectroscopy. Analyst 130, 1019-1026.
Jarvis, R.M. & Goodacre, R. (2004) Rapid discrimination of bacteria using surface enhanced Raman spectroscopy. Analytical Chemistry 76, 40-47.
López-Díez, E.C., Winder, C.L., Ashton, L., Currie, F. & Goodacre, R. (2005) Monitoring the mode of action of antibiotics using Raman spectroscopy: investigating sub-inhibitory effects of amikacin on Pseudomonas aeruginosa. Analytical Chemistry, 77, 2901-2906.
McGovern, A.C., Broadhurst, D., Taylor, J., Kaderbhai, N., Winson, M.K., Small, D.A., Rowland, J.J., Kell, D.B. & Goodacre, R. (2002) Monitoring of complex industrial bioprocesses for metabolite concentrations using modern spectroscopies and machine learning: application to gibberellic acid production. Biotechnology and Bioengineering 78, 527-538.
Maquelin, K., Kirschner, C., Choo-Smith, L.-P., van den Braak, N., Endtz, H. P., Naumann, D. & Puppels, G. J. (2002). Identification of medically relevant microorganisms by vibrational spectroscopy. Journal of Microbiological Methods 51, 255-271.
Shaw, A.D., Kaderbhai, N., Jones, A., Woodward, A.M., Goodacre, R., Rowland, J.J. & Kell, D.B. (1999) Non-invasive, on-line monitoring of the biotransformation by yeast of glucose to ethanol using dispersive Raman spectroscopy and chemometrics. Applied Spectroscopy 53, 1419-1428.




