Identification and prevalidation of safety biomarkers
Posted: 24 March 2006 | | No comments yet
To date, hazard/risk assessment of new drugs and chemicals primarily relies on the investigation of toxicological endpoints from animal studies. In this field, the full range of genomics and proteomics technologies can be used in efforts to uncover the molecular mechanisms at work in response to xenobiotic exposure. These new disciplines, called toxicogenomics and toxicoproteomics, […]
To date, hazard/risk assessment of new drugs and chemicals primarily relies on the investigation of toxicological endpoints from animal studies. In this field, the full range of genomics and proteomics technologies can be used in efforts to uncover the molecular mechanisms at work in response to xenobiotic exposure. These new disciplines, called toxicogenomics and toxicoproteomics, offer several practical benefits.
Due to a parallel approach, it should be possible to screen for toxic effects more rapidly than with conventional methods, such as histopathology and clinical chemistry/haematology. Since molecular changes occur prior to pathological outcomes, detection of disease and organ toxicity should be possible at earlier time-points during a pathological process. In addition, these technologies are highly sensitive, so that long-term toxic effects can be potentially detected at lower doses
Omics applications can be divided into two broad and partly overlapping classes – investigative studies and predictive toxicology. Investigative studies have been reported on new molecular targets for toxicants or gaining deeper insights into mechanisms of action (Man et al., 2002; Ruepp et al., 2002; Wetmore and Merrick, 2004; Fella et al., 2005; Hewitt et al., 2005). The belief that different groups or classes of compounds will induce specific genes/proteins or expression patterns provides the basis for predictive toxicology. Several publications show that such safety biomarkers/patterns can have a high degree of predictive power (Bandara, 2002; Elcombe et al., 2002; Li et al., 2002; Petricoin et al., 2002; Ellinger-Ziegelbauer et al., 2004, Ezendam et al., 2002). Described examples are promising to revolutionise the classical toxicology approaches that mainly investigate morphological changes by histopathology combined with clinical chemistry/haematology. However, in contrast to clinical investigations, toxicological studies are comprised of small sample sizes leading to the difficulty in confirming toxicology related Omics data. For this reason, genomics and proteomics methods, today, are mainly used for discovery approaches, while prevalidation and validation procedures on generated data are generally missing.
Safety biomarkers in toxicoproteomics
In terms of exploring a cells’ or organisms’ physiology, as well as drug/chemical induced changes the analysis of protein expression is of great interest. Toxicoproteomics can be described as a ‘marriage’ of classical toxicology assays and the new discipline of differential expression profiling – typically the relative quantitation of proteins in two sets of samples. Those experiments rely on comparison of corresponding expression levels and subsequent identification of differentially expressed proteins. Applications have been described for mechanistic investigations (Cutler et al., 1999; Bandara et al., 2003) for the pathological classification of diseased tissue (Roberts et al., 2003; Kröger et al., 2004) and ultimately for the discovery of toxicity related biomarkers, so-called safety biomarkers (Petricoin et al., 2002; Veenstra et al., 2004). Omics technologies provide a means for predicting toxicity prior to classical toxicological endpoints. This is due to the fact that these methods can detect even the smallest changes at the molecular level preceding traditional morphological and clinical endpoints. In addition, information on the pathways leading to toxic effects, the molecular mechanism, which can be obtained from protein expression profiles, will be of great value in the assessment of the relevance of alterations seen in animals. Moreover, one of the most important advantages of this strategy is the possibility of finding safety biomarkers that indicate toxicity endpoints earlier and with fewer resources.
Potential protein safety biomarkers can be detected by a broad spectrum of methods. All of them follow similar principles. After treatment of laboratory animals/cells, parallel protein extraction and protein separation is performed. Changes in the proteome are then investigated by differential expression analysis with bioinformatics/biostatistics tools before the identification of proteins of interest (Anderson and Anderson, 1998; Ryan and Patterson, 2001; Schrattenholz, 2004). The most powerful protein separation technologies are liquid chromatography (LC) and two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). For protein identification there is a set of mass spectrometry (MS) methods with ever increasing accuracy and sensitivity. Global measurements of protein expression can be performed by various combinations of these key technologies (Wilkins et al., 1997; Liebler, 2002). This has the potential to greatly impact toxicology over the next few years, and to help in the risk/hazard assessment of new drugs and chemicals. However, due to complexity of the proteome, all of these methods allow the visualisation of subproteomes only. All proteomic platforms do not yet deliver the same broad information as in the genomic field. Indeed, the relationship between gene number and proteome size is far from simple when taking into account splice variants and posttranslational modifications of proteins. Furthermore, the concentration range of the proteome exceeds seven orders of magnitude (high dynamic range) (Wilkins et al., 1996; Harrison et al., 2002). Single proteomic platforms are not sufficient to display the enormous proteome in total. For this reason, only combinations of multiple protein analysis technologies will aid in the discovery and prevalidation of protein biomarkers in complex problem areas like toxicology.
Prevalidation of safety biomarkers
The discovery of potential safety biomarkers in modern toxicology results in a large panel of protein candidates. Due to a lack of specific antibodies, the prevalidation and validation of these proteins requires the implementation of new methods besides conventional ELISA or Western Blotting (WB) techniques. The developments of tandem mass spectrometry as well as the improvements of LC-technologies allow the detection of proteins with an increased sensitivity compared to 2D-PAGE/MS approaches. The detection of thousands of proteins from 104 cells is possible using LC-based methods (Chamrad and Meyer, 2005). In addition, new MS technologies that were originally used for the identification and quantification of small molecules have been recently applied to protein identification (Unwin et al., 2005). As the peptide structure and mass is known, this information can be used to design a workflow for the sequencing of targeted peptides out of complex peptide mixtures derived from intricate protein lysates like liver, kidney or plasma proteomes. This workflow can be easily applied to prevalidate potential marker protein candidates derived from classical proteomics approaches.
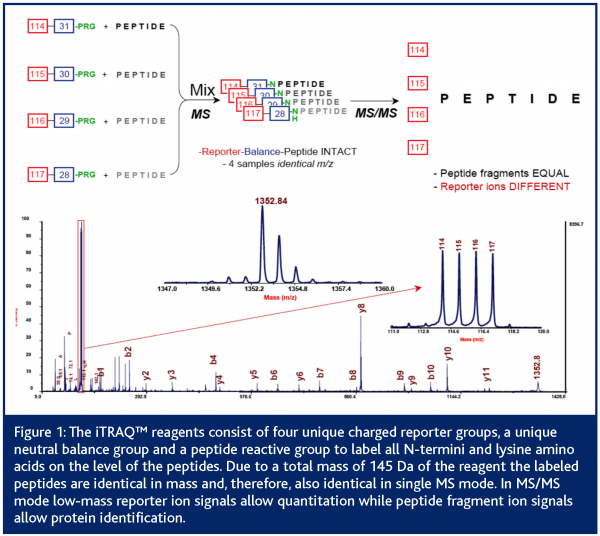
Besides some techniques that allow relative quantitation between different LC-MS runs (Schulz-Knappe et al., 2003) most LC-MS based approaches focus on the comparison of isotopically labelled peptide samples (Pan et al., 2005; Mann, 1999). Stable isotope tagging methods provide a useful means of determining the relative expression levels of individual proteins between samples (Lill, 2003; Schneider and Hall, 2005). First experiments for quantitative mass spectrometry (QMS) go back to R. Aebersold and co-workers (Gygi et al., 1999). They used isotope-coded affinity tags (ICAT) to chemically label cysteine residues in proteins before protein digestion and quantitation by MS. The ICAT® reagents contain a biotin affinity tag to allow the enrichment of cysteine-containing peptides with avidin affinity chromatography. An acid-cleavable linker is incorporated to allow the removal of the biotin affinity tag before MS or MS/MS analysis is performed. Since the development of ICAT® reagents, many groups have adopted this strategy and created several stable isotope-tagging reagents targeting different amino acids (Moritz et al. 2003; Schmidt et al. 2005). The latest development for chemical labelling is the isotope tag for relative and absolute quantitation (iTRAQ) (Ross et al., 2004). These reagents target all N-termini as well as lysine residues of the peptides before MS/MS based quantitation can be performed. The iTRAQ™ reagent methodology utilises isobaric tags containing a charged reporter group and a second balance group. While labelled peptides are identical in single MS mode, the unique reporter group (masses between 114 and 117 Da) allows quantitation and protein identification of peptides in MS/MS mode (Figure1).
While neither 2DE-PAGE/MS, LC/MS approaches or isotope tagging methods are able to cover the full range of the proteome, the latter shows the clear advantage of automation and reproducibility. Due to the high sensitivity and novel workflows, this quantitative mass spectrometry (QMS) technology is gaining importance in the field of proteomics. QMS is useful both for investigative studies and with regard to discovery of new safety markers for predictive toxicology. Furthermore, this approach can be adapted for prevalidation procedures of safety biomarkers.
Toxicoproteomics at Merck – Identification and prevalidation of safety biomarkers predicting carcinogenicity
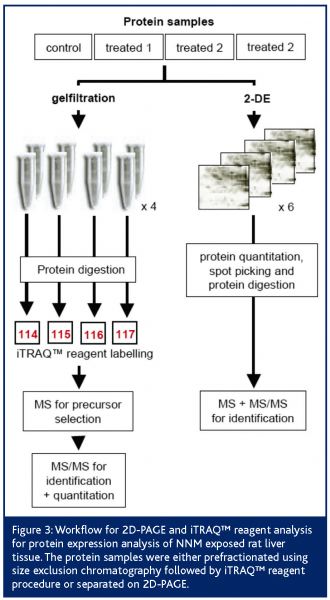
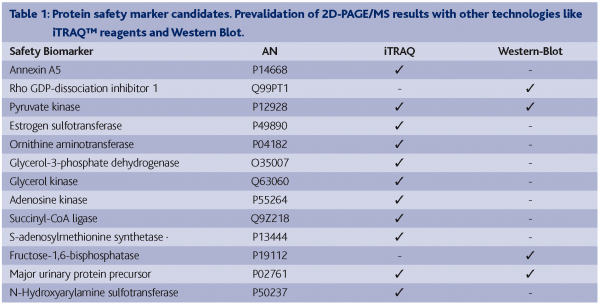
A common animal model of chemical hepatocarcinogenesis was successfully used to demonstrate the early detection of safety biomarkers related to the classical toxicological endpoint, cancer. Liver carcinoma and adenoma were induced in male Wistar rats after 25 weeks by treating them with the well characterised liver carcinogen N-Nitrosomorpholine (NNM). By using the 2D-PAGE approach in combination with MS based protein identification, we were able to identify statistically significantly deregulated proteins in NNM exposed rat liver tissue in late as well as early time-points (Table 1). These latter proteins might have predictive power for the chemically induced liver carcinogenicity in rats. Biological relevance of these candidate early safety biomarkers has been proven (Fella et al., 2005). However, in order to ensure reliable results, independent technology platforms have to be used to prevalidate these protein expression patterns, for example:
- MS based protein profiling approaches
- Antibody based technologies
- Gene expression profiling technologies
- Quantitative mass spectrometry
With the aim to prevalidate the potential 2D-PAGE/MS derived candidate early markers, antibody based technologies such as Western Blotting (WB) have been applied. However, this approach was not completely successful as only a few of all tested antibodies showed satisfying specificity and sensitivity, and in fact, most of the desired antibodies were not commercially available. Nevertheless, down regulation of the proteins α2μ-globulin (major urinary protein precursor), pyruvate dehydrogenase kinase 1 (pyruvate kinase) and fructose-1,6-bisphosphatase and the up regulation of Rho-GDI dissociation inhibitor could be confirmed by using this technique (Table 1).
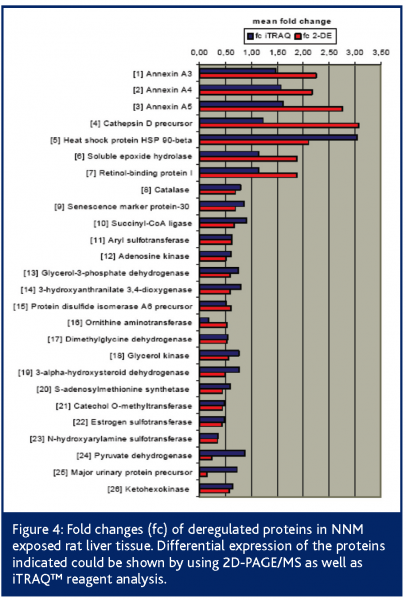
Additionally, we used the iTRAQ™ reagent technology for prevalidation. Following the peptide labelling and MS/MS based identification and quantitation; the regulation of 26 2D-PAGE/MS derived proteins could be confirmed at all (Figure 3). Proteins like heat shock protein 90-beta, annexin A5, ketohexokinase, N-hydroxyarylamine sulfotransferase, ornithine aminotransferase and adenosine kinase showed highly comparable fold changes using both proteomic quantitation strategies (Figure 4). Furthermore, 11 of the potential early safety biomarkers, which might have predictive power for chemically induced liver carcinogenicity, were prevalidated (Table 1). In addition, iTRAQ™ reagent analysis delivered further potential safety biomarkers with biological relevance to the processes of hepatocarcinogenicity, e.g., the placental form of glutathione S-transferase (GST-P), carbonic anhydrase and aflatoxin B1 aldehyde reductase. These results show the power of this new quantitation strategy for the verification of candidate safety biomarkers in addition to the discovery of new potential safety biomarkers.
Conclusions
The prevalidation of safety biomarker candidates in toxicology requires the application of new methods besides antibody-based technologies like ELISA or Western Blotting. Due to the known poor correlation between protein and gene expression, genomics technologies seem to be questionable for prevalidation procedures of protein biomarkers. Nevertheless, new mass spectrometry based technologies allow reliable quantitation of proteins. Our results show the usefulness of the iTRAQ™ reagent technology for biomarker prevalidation on one hand, as well as for identification of new potential safety marker proteins, indicative of hepatocarcinogenicity, on the other hand – combined with a significant decrease in instrumental and working time compared to classical combination of 2D-PAGE with mass spectrometry based protein identification and quantitation.






References
Anderson NG, Anderson NL (1998) Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 19: 1853-1861
Asara JM, Zhang X, Zheng B, Christofk HH, Wu N, Cantley LC (2006) In-Gel Stable-Isotope Labeling (ISIL): A Strategy for Mass Spectrometry-Based Relative Quantification. J. Proteome Res. 5: 155-163
Bandara LR, Kennedy S (2002) Toxicoproteomics – a new preclinical tool. Drug Discovery Today 7: 411-418.
Bandara LR, Kelly MD, Lock EA, Kennedy S (2003) A correlation between a proteomic evaluation and conventional measurements in the assessment of renal proximal tubular toxicity. Toxicol. Sci. 73: 195-206
Chamrad D, Meyer HE (2005) Valid data from large-scale proteomics studies
Nature Methods 2, 647-648, News and Views
Cutler P, Bell DJ, Birrell HC, Connelly JC, Connor SC, Holmes E, Mitchell BC, Monte SY, Neville BA, Pickford R, Polley S, Schneider K, Skehel JM (1999) An integrated proteomic approach to studying glomerular nephrotoxicity. Electrophoresis 20: 3647-3658
Elcombe CR, Odum J, Foster JR, Stone S, Hasmall S, Soames AR, Kimber I Ashby J (2002) Prediction of rodent nongenotoxic carcinogenesis: evaluation of biochemical and tissue changes in rodents following exposure to nine nongenotoxic NTP carcinogens. Environ Health Perspect. 110: 363-375
Ellinger-Ziegelbauer H, Stuart B, Wahle B, Bomann W, Ahr HJ (2004) Characteristic expression profiles induced by genotoxic carcinogens in rat liver. Tox Science 77: 19-34
Ezendam J, Staedtler F, Pennings J, Vandebriel RJ, Pieters R, Boffetta P, Harleman JH, Vos JG (2004) Toxicogenomics of subchronic hexachlorobenzene exposure in Brown Norway rats. Environ Health Perspect. 112(7): 782-791
Fella K, Glückmann M, Hellmann J, Karas M, Kramer P-J, Kröger M (2005) Use of two-dimensional gel-electrophoresis in predictive toxicology: Identification of potential early protein biomarkers in chemically induced hepatocarcinogenesis. Proteomics 5: 1914-1924
Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology 17: 994-999
Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M (2002) A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 30: 1083-1090
Hewitt P, Kramer P-J, Borlak J (2005) Toxicogenomics applied to teratogenicity studies. In: Borlak J (ed) Handbook of Toxicogenomics, Strategies and Applications. Wiley-VCH, Weinheim, pp 435-469
Kröger M, Hellmann J, Toldo L, Glückmann M, von Eiff B, Fella K, Kramer PJ (2004). Toxicoproteomics: First experience in a BMBF study. Altex 21: 28-40
Kennedy S (2002) The role of proteomics in toxicology: identification of biomarkers of toxicity by protein expression analysis. Biomarkers 7: 269-290
Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW (2002) Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clinical Chemistry 48: 1296-1304
Man WJ, White IR, Bryant D, Bugelski P, Camilleri P, Cutler P, Heald G, Lord PG, Wood J, Kramer K (2002) Protein expression analysis of drug-mediated hepatotoxicity in Sprague-Dawley rat. Proteomics 2:1577-1585
Mann M (1999) Quantitative proteomics? Nature Biotechnology 17, 954-955
Lill J (2003) Proteomic tools for quantitation by mass spectrometry. Mass Spectrometry Reviews 22: 182-194
Moritz B, Meyer HE (2003) Approaches for the quantification of protein concentration ratios. Proteomics 3: 2208–2220
Pan S, Zhang H, Rush J, Eng J, Zhang N, Patterson D, Comb MJ, Aebersold R (2005) High Throughput Proteome Screening for Biomarker Detection.Mol. Cell. Proteomics 4: 182-190
Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC and Liotta LA (2002) Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359: 572-577
Roberts R, Cain K, Coyle B, Freathy C, Leonard JF, Gautier JC (2003) Early drug safety evaluation: biomarkers, signatures and fingerprints. Drug Metab Rev. 35: 269-275
Ruepp SU, Tonge RP, Shaw J, Wallis N, Prognan F (2002) Genomics and proteomics analysis of acetaminophen toxicity in mouse liver. Toxicol Sci. 65: 135-150
Ryan TE, Patterson SD (2001) Proteomics in drug target discovery – high-throughput meets high-efficiency. Drug Discovery World 2: 43-52
Schmidt A, Kellermann J, Lottspeich F (2005) A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 5: 4–15
Schneider LV, Hall MP (2005) Stable isotope methods for high-precision proteomics. Drug Discovery Today 10: 353-363
Schrattenholz A (2004) Proteomics: how to control highly dynamic patterns of millions of molecules and interpret changes correctly. Drug Discovery Today: Technologies 1: 1-8
Schulz-Knappe P, Schrader M (2003) Peptidomics in biomarker and drug discovery. Current Drug Discovery 21-24
Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M (2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. PROTEOMICS 3: 377-396
Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD (2005) Multiple Reaction Monitoring to Identify Sites of Protein Phosphorylation with High Sensitivity. Mol. Cell. Proteomics 4: 1134 – 1144
Veenstra TD, Prieto DA, Conrads TP (2004) Proteomic patterns for early cancer detection. Drug Discovery Today 9: 889-897
Wetmore BA, Merrick BA (2004) Toxicoproteomics: Proteomics applied to toxicology and pathology. Toxicologic Pathology 32: 619–642
Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humpherey-Smith I, Hochstrasser DF, Williams KL (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnology and Genetic Engineering Reviews 13: 19-50




