Quantitative PCR assays in clinical drug development
Posted: 28 September 2006 | | No comments yet
Biomarkers (biological markers) have become an integral part of both drug discovery and drug development and play an important role in the transition of potential new drugs from discovery into clinical drug development. In the past, most biomarkers were proteins/peptides and metabolites measured by technologies such as immunoassays, enzymatic assays, HPLC and mass spectrometry.
Biomarkers (biological markers) have become an integral part of both drug discovery and drug development and play an important role in the transition of potential new drugs from discovery into clinical drug development. In the past, most biomarkers were proteins/peptides and metabolites measured by technologies such as immunoassays, enzymatic assays, HPLC and mass spectrometry.
Biomarkers (biological markers) have become an integral part of both drug discovery and drug development and play an important role in the transition of potential new drugs from discovery into clinical drug development. In the past, most biomarkers were proteins/peptides and metabolites measured by technologies such as immunoassays, enzymatic assays, HPLC and mass spectrometry.
The introduction of PCR-based technologies during the early 1990s, the decoding of the human genome in the late 1990s and the proliferation of microarray-based experimentation during the last decade have added a whole new set of DNA- and RNA-based biomarkers for potential use in drug discovery and development. Quantitative methods for assessing RNA levels must be analytically validated to ensure that data generated will be of high quality and provide meaningful quantitative information about the levels of the transcriptional biomarker in a clinical study. The importance of molecular biology and the rapid growth of new molecular technologies in medicine is evidenced by the recent change in the newest edition of the Tietz Textbook of Clinical Chemistry to the Tietz Textbook of Clinical Chemistry and Molecular Diagnostics.1
Biomarkers in clinical drug development
Pharmaceutical companies are using biomarkers in early clinical drug development (phase I) to enable early proof-of-concept studies and to better predict the dose range for the larger phase II and III studies.2 Together with the pharmacokinetic (PK) data, the pharmacodynamic (PD) biomarker data can be used to demonstrate a relationship between drug concentrations and drug effects or side effects as measured by the PD biomarker(s). The PK/PD relationship can support internal business decision-making regarding dose and dose regimen.
Very few biomarkers e.g. HIV plasma viral load and cholesterol, have reached the level of surrogate endpoints substituting for clinical endpoints.3 There is currently no regulatory guidance for the development and validation of methods used to measure research-type (non-diagnostic) biomarkers and most PD biomarkers belong to this class. Cross-functional teams will define the intended use of a biomarker or a set of biomarkers in clinical drug development and this will, to a great extent, dictate the validation of the biomarkers and the associated methods.4 Many pharmaceutical companies have internal biomarker assay validation committees that will review and give final approval to use biomarker assays in clinical drug development based on the reported analytical performance of the assays.
Analytical method validation
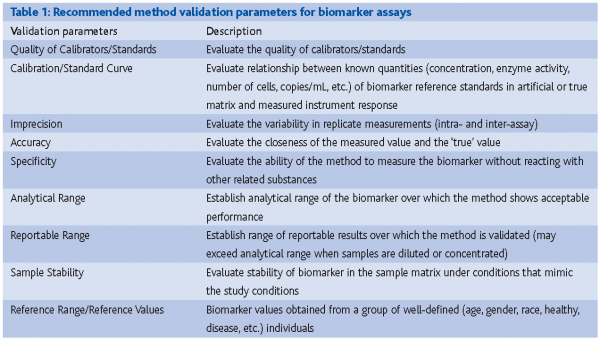
Most published articles outlining validation parameters (e.g., precision, accuracy, and concentration-response relationship) for quantitative methods have focused on method validation in support of measurements of drugs using mass spectrometry, HPLC and immunoassay based technologies.5,6 Analytical laboratories involved with measurements of biomarkers are familiar with these validation parameters.4 However, method validation for biomarker assays tends to be more complicated than that for drug assays and it is often not possible to evaluate all of the validation parameters outlined in the FDA guidance for Method Validation when validating biomarker assays.7 It is therefore up to the analytical laboratories to set their own recommendations and standards for the performance assessment of biomarker methods (Table 1).
The main objective of method validation is to characterise and control sources of variability. Often too much emphasis is given to pre-specified method performance criteria. For example, a PD biomarker method that shows an imprecision of 30-40% CV at low concentrations of the biomarker measured in specimens collected prior to dosing can be acceptable if the effect of the drug leads to a 5-fold increase in the concentrations and at the elevated concentrations, the imprecision of the assay is 5-15% CV. Determination of the absolute accuracy of a biomarker assay can be very challenging if well-characterised reference materials/standards or reference methods are not available.
To control for changes in method performance, laboratories must use quality control (QC) samples. The QC samples should, whenever possible, be based on the true matrix and acceptance criteria (usually mean ±2SD or the 95% confidence interval for quantitative assays) determined based on the actual inter-assay variability in the laboratory performing the assay for the clinical studies. By depicting the performance of the QC samples during a clinical study, a more accurate picture of the actual assay performance during the study can be determined.
Quantitation options for real-time RT-PCR methods
There is an acknowledged need to apply quantitative approaches to confirm and extend initial microarray-based observations with alternative technologies that offer higher degrees of quantitative accuracy. Although a number of RNA quantitation methodologies exist (real-time RT-PCR, branched DNA amplification, etc.), the present review focuses on the widespread real-time RT-PCR methodology.8 Even within real-time RT-PCR methods there are several different fluorescence detection strategies and many real-time RT-PCR approaches are available.9-11
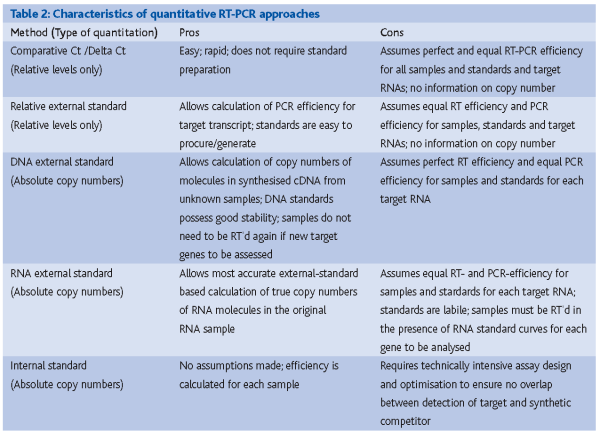
Currently a large number of quantitative approaches exist for real-time PCR including 1) comparative cycle threshold (Ct) approaches, 2) relative quantitative approaches, 3) external standard approaches and 5) internal-standard approaches. A table describing the quantitative characteristics and pros and cons of each approach is presented in Table 2.
The comparative Ct approach involves determining the difference between the cycle threshold (Ct) for a target gene in one sample and the Ct for that target gene in a second sample (termed the “delta” Ct, or ΔCt). If a normalisation gene is used, the difference in the Ct between the target and the reference gene represents the ΔCt in sample one and a second ΔCT is calculated for the difference in Ct between the target and reference gene in sample two. Since PCR amplification is exponential, the fold-difference between the normalised levels of the target gene in two samples is approximated by the following formula:
Fold difference = 2 ^ (ΔCtsample 1 – ΔCtsample 2)
The comparative-Ct method assumes 100% efficiency for PCR amplification, which is rarely the case and the methods are incapable of giving any information about actual copy numbers of the target transcripts measured. We have previously used the delta-delta Ct approach to confirm microarray signatures identified in clinical pharmacogenomic studies in which expression profiles were generated during the clinical study.12 However, it is unclear whether the quantitation afforded by this approach will suffice for the accurate quantitation required for pharmacodynamic measurements of transcriptional biomarkers in early clinical studies.
Relative external standards involve the dilution of a total RNA sample of unknown starting copy number and the derivation of a standard curve relating measured Ct values for the target gene to the dilution series. Although this method is more robust than a comparative Ct approach because it allows the efficiency of PCR amplification to be calculated, it is limited by the assumption that efficiency is identical between standards and samples and because it provides no information on the actual copy number present in the samples.
External standard and internal standard approaches represent the most quantitative approaches available in quantitative RT-PCR (qRT-PCR) today and enable users to calculate copy numbers of target transcripts if the appropriate standards are used. External standard approaches are typically easier to execute and very robust but nonetheless still make the assumption inherent to all external standard approaches, namely that amplification efficiency is identical between the external standards and the unknown samples. It should be noted that RNA-, rather than DNA-, external standard approaches are the most robust because they provide the user with an accurate estimate of both reverse transcription and PCR efficiency, since DNA-based approaches only capture PCR efficiency. RNA standards, on the other hand, are far less stable and more difficult to work with than the corresponding cDNA standards. In addition, each new gene analysed in a set of samples by an RNA standard method will require a new round of reverse transcription of the standard curve along with the samples, whereas cDNA standard curves can simply be added alongside previously reverse transcribed samples and subjected to PCR.
Finally, internal standards represent the most stringent quantitation approach because this is the only method whereby the amplification efficiency within each sample can be calculated.13 This is an especially important consideration if PCR inhibitors may be variably present in the clinical sample matrix to be analysed.
Analytical validation of qRT-PCR assays in clinical drug development
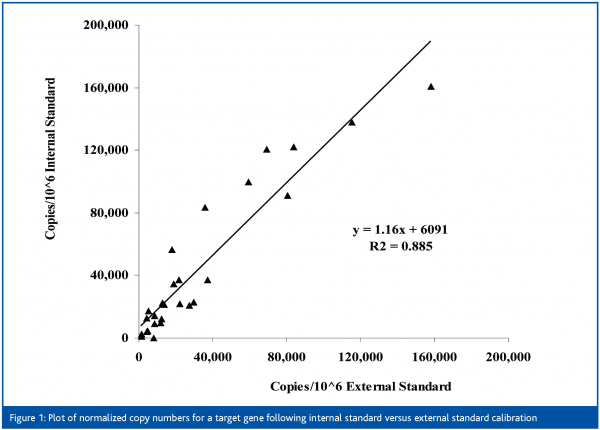
We have evaluated all of the above approaches in the past and depending on the sample type and the clinical question of interest, RNA external standard and/or internal standard approaches appear to give the most robust results. Figure 1 demonstrates the actual correlation between normalised copy numbers calculated from internal and external standard approaches for a target gene. Because of space limitations, the steps involved in the analytical validation of an external standard assay are described in the next section. However, all (or most) of the analytical performance principles described can be applied to qRT-PCR approaches in general.
The first step in analytical validation involves the generation of high quality reagents for calibration curves. This is typically performed by in vitro transcription of the RNA target from a cDNA within a linearised cloning vector. Highly purified, uniform-length standards are preferred to ensure that quantitation estimates of the in vitro transcription reaction product by absorbance accurately reflect the concentration of the full-length transcript. The copy numbers are calculated based upon the known size of the in vitro transcript.
The next step is the selection of the reference or normalisation gene. We typically screen a panel of normalisation genes prior to reference gene selection. Factors influencing the selection of the reference gene include assessment of its variability across multiple healthy subjects, its stability and reproducibility during the sample collection method, and the non-inducibility of the reference gene following direct exposure to the drug candidate to be tested. In some instances the last factor can be assessed by treatment with the compound in ex vivo culture.
Calibration curves are generated by spiking known amounts of the purified standards in replicates at 6-10 concentrations following 10-fold serial dilutions. The intra- and inter-assay performance of the calibration curve is determined in five or more analytical runs. For the intra-assay imprecision the mean, SD, imprecision (expressed as the coefficient of variation, %CV), and the inaccuracy (expressed as the difference from the ‘true’ copy number, % bias) of the calculated concentrations are calculated for each replicate in each analytical run of the calibration curves. Additionally, the inter-assay imprecision is calculated for all analytical runs.
In real-time PCR assays we have defined the lower limit of quantitation (LLQ) as the lowest copy number that can be measured with an acceptable imprecision (%CV) and % bias (an imprecision of ±30% and a bias of ±30% are generally considered acceptable for qRT-PCR assays). The LLQ should also represent a copy number level that generates Ct values < 36 cycles. The upper limit of quantitation (ULQ) is the highest concentration measured with an acceptable (see above) %CV and % bias. The interval between the LLQ and the ULQ is the quantitative concentration range of the qRT-PCR calibration curve.
After the performance of the calibration curve is established, validation samples are prepared by spiking transcripts into total RNA samples to evaluate the analytical performance of the assay using samples of at least three divergent concentrations (low-range, mid-range and high-range). In the best case scenario these validation samples are prepared in large enough quantities that they can also become the QC samples used during the analysis of clinical samples, whenever possible. The imprecision of the qRT-PCR assay during the analysis of clinical samples will be monitored through the QC sample results. At least two aliquots of each of the three QC samples (QC-low, QC-mid, QC-high) are analysed in each analytical run. Acceptance ranges (mean ± 2SD) are typically established by determining the inter-assay imprecision of each QC sample.
The accuracy is defined as the closeness of the measured value of an analyte to its ‘true’ value. The ‘true’ value of an analyte in a biological matrix is usually established using a reference method. However, for many biomarkers there are no reference methods available, in which case accuracy is usually assessed by measuring the recovery of the analyte when added (spiked) to the biological matrix.
The specificity of the assay relates to the ability of the assay to recognise and accurately measure the specified analyte without reacting with or detecting other related substances. Information about the specificity of the PCR primers used in any given qRT-PCR assay can typically be predicted by bioinformatic analysis of blast search results against the most recent build of the human genome database. The extensive bioinformatics process for PCR primer and probe design used in commercially available assays-on-demand (Applied Biosystems) is detailed in a white paper and indicates the complexity well.14 However, the specificity of a PCR-based assay can also be demonstrated, rather than predicted in silico, by a variety of approaches including melting curve analysis or RFLP or DNA sequencing of the isolated PCR product.
The short-term stability of the transcript in the biological matrix should be evaluated at the temperatures under which storage and isolation will be performed. Experiments will assess the stability of the analyte in the biological matrix (when the matrix is prepared from whole blood, the stability in whole blood should be evaluated) during the collection, processing, handling and storage of the sample. Both short- and long-term stability should be assessed by measuring the endogenous analyte concentrations in the biological matrix from several individual subjects. The freeze-thaw stability (up to three F/T cycles) of isolated total RNA from several individual subjects should also be assessed.
Whenever possible, the reference range/reference values (the use of ‘normal range’ should be avoided) of the transcript in apparently healthy subjects should be determined by measuring the transcript concentration in samples collected from preferably 40 or more subjects (detailed descriptive statistics of the subjects should be given e.g. age, gender, ethnicity, etc.). Particularly in preparation for first-in-human studies, knowledge of the anticipated levels of the target in apparently healthy individuals is of importance to evaluate whether the calibration range of the assay will suffice for the clinical study.
Conclusion
In conclusion, the precepts of analytical validation for biomarker assays have been established over many years. The implementation of microarray technologies during the drug discovery process to identify potentially robust and reliable biomarkers will continue to drive the need for analytical validation of qRT-PCR assays capable of measuring transcriptional biomarker levels accurately in clinically accessible matrices. While many of the concepts for analytical validation of molecular diagnostic assays can be applied to the analytical validation of qRT-PCR assays used for drug development, many drug-specific issues must also be addressed. With careful planning and patient execution of analytical method validation studies prior to implementation of assays in the analysis of clinical samples, high quality measurements of transcriptional biomarkers will be able to enhance the development of pharmaceuticals in the future.



References
- Tietz Textbook of Clinical Chemistry and Molecular Diagnostics. Ed. Carl A. Burtis, Edward R. Ashwood, and David E. Bruns. Elsevier Saunders, St. Louis, MO
- Bjornsson TD. Applications in drug development. European Pharmaceutical Review 10(1), 17-21 (2005)
- Biomarker Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther 2001; 69:89-95.
- Swanson B. Delivery of high-quality biomarker assays. Disease Markers 2002; 18:47-56.
- Shah VP, Midha KK, Findlay JWA, et al. Bioanalytical method validation?A revisit with a decade of progress. Pharm Res 2000; 12:1551-1557.
- DeSilva B, Smith W, Wiener R, et al. Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules. Pharm Res 2003; 20:1885-1900.
- Guidance for Industry, Exposure-Response Relationship – Study Design, Data Analysis, and Regulatory Applications. U.S. Department of Health and Human Services, Food and Drug Administration, April 2003.
- Higuchi R, Fockler C, Dollinger G and Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology 1993; 11: 1026-1030.
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endo 2000; 25: 169-193.
- Wilhelm J and Pingoud A. Real-time polymerase chain reaction. ChemBioChem 2003; 4: 1120-1128.
- Wong ML and Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques 2005; 39: 75-85.
- Burczynski ME, Peterson RL, Twine N, Zuberek K, Brodeur BJ, Casciotti L, Spinelli W, Cotreau MM, Schwertschlag U, Immermann F, and Dorner AJ. Molecular classification of Crohn’s disease and ulcerative colitis patients by global expression analysis of peripheral blood mononuclear cells. J Mol Diag. 2006; 8: 51-61.
- Crawford EL, Warner KA, Khuder SA, Zahorchak RJ, and Willey JC. Multiplex standardized RT-PCR for expression analysis of many genes in small samples. Biochem Biophys Res Commun. 2002; 293:509-516.
- Lazaruk K, Wang Y, Zhong J, Maltchenko S, Rabkin S, Hunkapiller K, Furtado M, Petrauskene O, Guegler K, Gilbert D and Spier E. The design process for a new generation of quantitative gene expression analysis tools: TaqMan probe-based assays for human, mouse and rat genes. www.appliedbiosystems.com (Accessed August 27, 2006)





