Cleanroom microbiology: single-temperature incubation for EM
Posted: 8 July 2024 | Guillaume Pinon (Servier) | No comments yet
Single-temperature incubation has been discussed for a long time in the pharmaceutical industry but only a few sites have implemented this system. Here, Guillaume Pinon, Head of the Microbiology Lab at Servier, discusses the use of single-temperature incubation in an aseptic production facility, outlining the regulatory considerations, strategic approach, challenges and prospects.

Single- or mono-temperature incubation is not easy to implement, but it offers efficiency and performance advantages and an opportunity to simplify the process in a pharmaceutical microbiology laboratory. This article shares our motivations, approach and experience of implementing mono-temperature incubation in our laboratories over the past several years.
Regulatory requirements
First we looked at the regulatory texts available concerning mono-temperature incubation for environmental monitoring (EM). The search was straightforward, as there is no description in pharmacopoeias or official regulatory guidance. Information can be found in certain US pharmacopeia guide chapters, such as USP <1116>,1 as well as guidance from the US Food and Drug Administration (FDA) and World Health Organization (WHO) but none of these are regulatory requirements. From this observation, all options are possible. However, adoption is not simple and one point frequently made is the need to take historical data into account. In the absence of any regulatory text, numerous presentations have been made on the subject, prompting debate and initiating reflection.2,3 Mono-temperature incubation will also receive special attention when A3P Association’s Microbiology common interest group (CIG) members carry out specific work.
Study: Strategy
To understand our choice of mono-temperature incubation, several factors need to be taken into consideration.
The first factor is our need to develop an environmental monitoring plan for a new unit for bioproduction of monoclonal antibodies and aseptic filling under isolator. The unit comprises 2,000m2 of Grade C and D, as well as an isolator for aseptic filling.
This information eliminates the need for comparison with historical data, enabling a simpler approach to implementation. The method described below should not be discarded, however, but adapted for comparison with historical data obtained with a double-temperature incubation sequence.
Cleanroom infrastructure innovation to fulfil the needs of cell and gene therapies
The second factor is a desire to simplify and optimise the process of handling environmental controls.
These two reasons prompted us to launch a wide-ranging study comparing incubation sequences involving a multitude of strains, in order to produce sufficient data to support a solid argument for the selection of an incubation sequence.
Step 1: Culture medium selection
This first step is essential, as the selected medium must enable the recovery of multiple strains from different ecosystems (eg, human, environmental) and of different natures (eg, bacteria, yeasts, moulds). We chose the tryptone soy agar (TSA) with neutralisers medium, which allows most microorganisms to thrive thanks to its nutrient conditions.
As taxonomy is extremely diverse, varied and not yet fully understood, it is undeniable that strains do exist which are not able to grow on this medium, but it does have the nutrients required for the development of strains commonly found in a pharmaceutical environment.
Step 2: Strain selection
Strain selection was launched in two test phases.
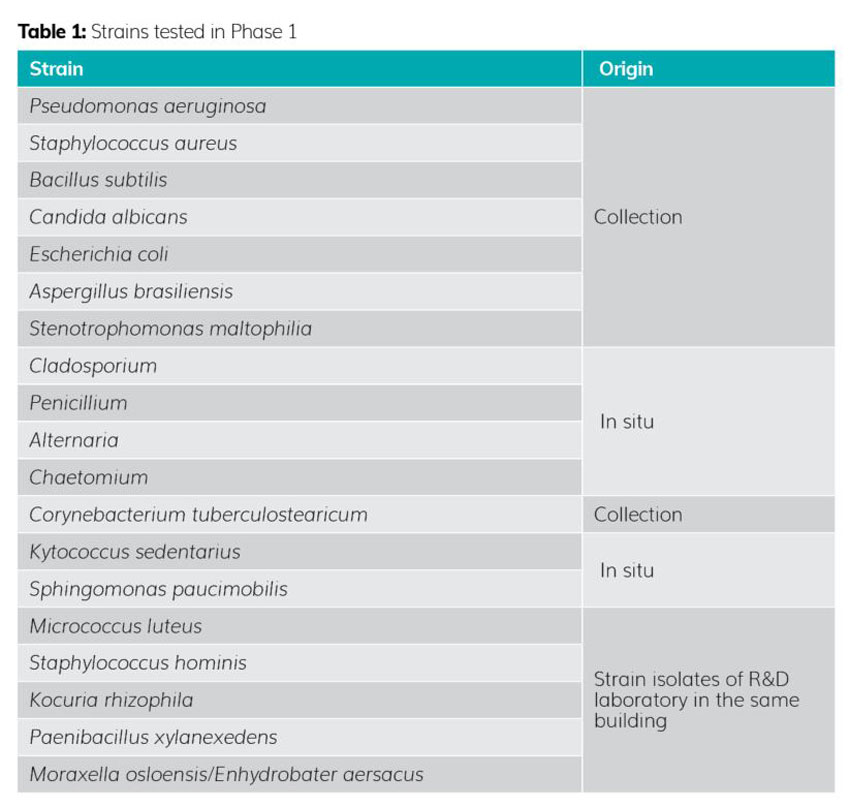
Phase 1 involved testing 19 strains (see Table 1). Each strain was selected to take into account:
- the variability of environmental and human strains in a pharmaceutical production environment
- strains known for their particular growth conditions, such as growth temperature, invasiveness, growth difficulties, etc (eg, Cladosporium, specific moulds, corynebacteria, Bacillus, etc).

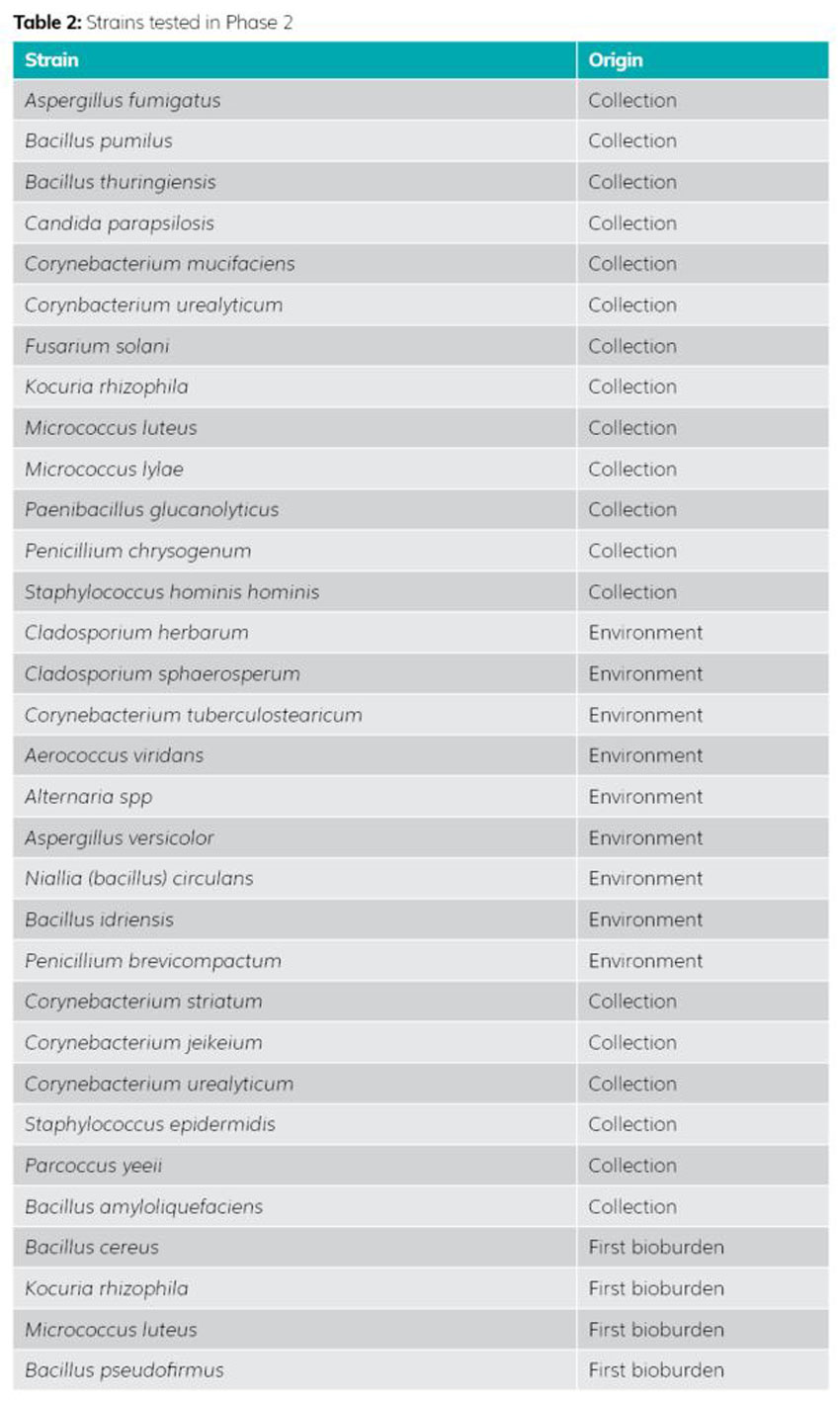
Phase 2 included selection of 32 new strains mainly found in the pharmaceutical industry (see Table 2). This identification was carried out in partnership with the ACM PHARMA laboratory, which performs around 35,000 identifications per year, giving us a more complete panel of different species generally encountered in the pharmaceutical industry. This selection completes the large families of strains previously tested in Phase 1. For example, a strain of Cladosporium was tested in the first phase because it was known not to grow at temperatures above 26-27°C (the worst-case strain in the study). In the second phase, two other Cladosporium strains were added to confirm the results generated. A Corynebacterium strain was also tested, as it has a growth preference for high temperatures. One species was tested in Phase 1 and supplemented by six new species in Phase 2. This multiplication of species and strains in the second phase ensures the reproducibility of the results generated for a given temperature and strain typology.

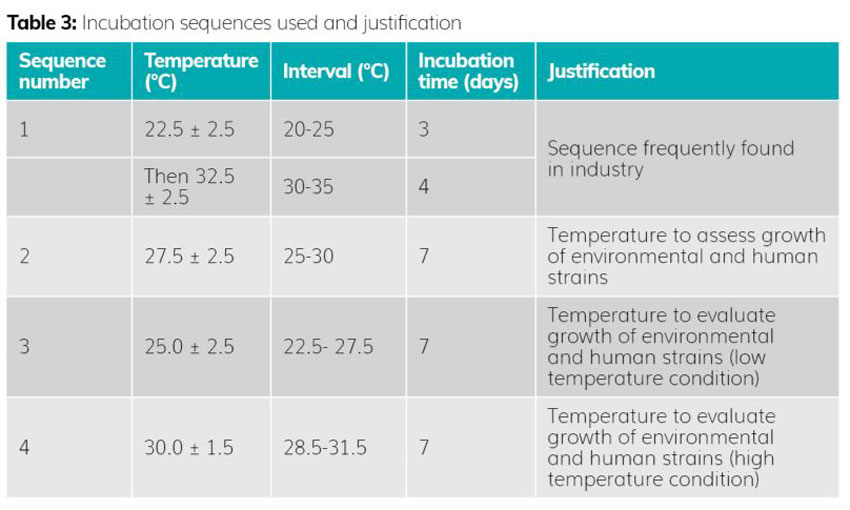
Step 3: Selection of incubation sequences
The selection of incubation temperatures was based on the data shown in Table 3.
The whole of Phase 1 (19 strains) was carried out under standard laboratory conditions, ie, incubation in ovens and manual enumeration.

In Phase 2 (addition of 32 new strains), we augmented the standard conditions with the use of an automated system for incubation and real-time reading. The system was set up for incubation at the target temperature of 25°C (based on the results of the first test phase). This new entrant provides data for an essential EM parameter: early detection of microbial growth. This element is essential for reactivity in the transmission and treatment of a contamination event in a classified area.
Results
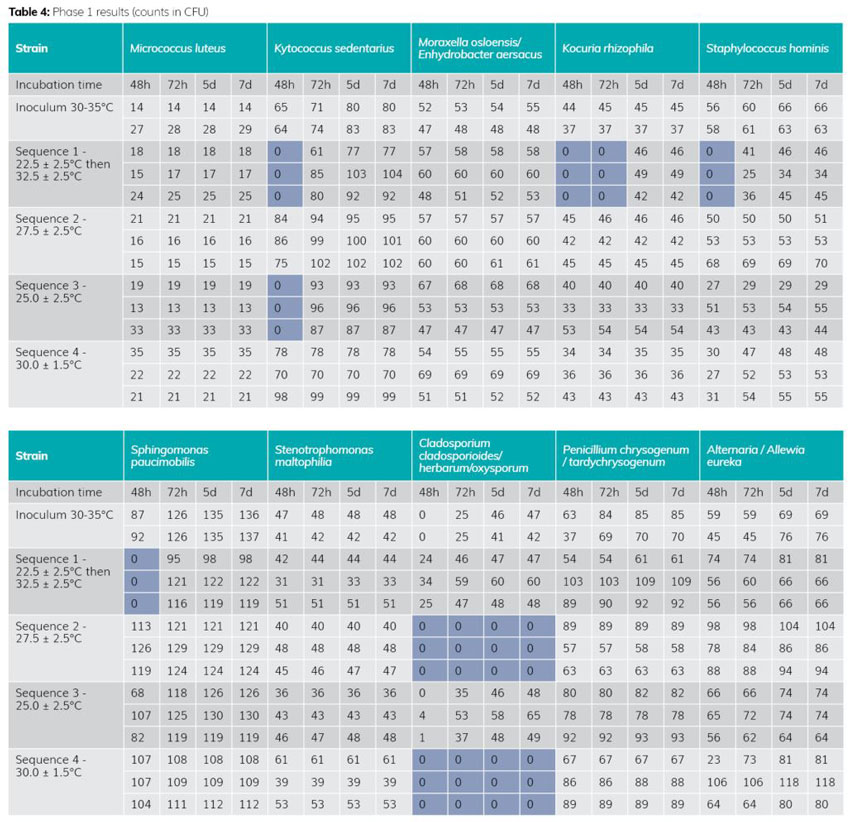
The results of Phase 1 (see Table 4) highlight 25°C as the most effective temperature for recovery. In fact, this temperature does not inhibit growth of the Cladosporium strain, unlike 27.5°C and 30°C.
The double-temperature sequence produces less effective results in the first 48-72 hours on strains from the cockle family (one of the majority families in a classified area). This information is important, as the slower growth induced by the lower temperature at the start of incubation could, on the one hand, be a source of inhibition for fragile microorganisms and, on the other, be less compatible with early detection of contamination in the classified area.
The 25°C temperature is therefore the most effective, as it does not inhibit any strain (compared with temperatures of 27.5°C and 30°C), and of the panel tested, only one strain develops slowly (no visible colonies) in the first 48-72 hours (instead of four strains for the double temperature).
Phase 1 demonstrated that the 25°C temperature performed best, of the sequences tested, and that it was suitable for the detection of environmental microorganisms. However, the panel of strains tested was not sufficiently complete, prompting the move to Phase 2 of the study.

The second phase is still in progress on moulds and some bacteria, with 39 strains tested out of the 51. The results of Phase 2 so far show no significant difference in the behaviour of the additional strains. Each family reacted in the same way (corynebacteria, Staphylococcus, Micrococcus, Bacillus, etc). The new strains tested (eg, paracoccus yeii or paenibacillus glucanolyticus) confirm the temperature of 25°C.
These results therefore confirm that the 25°C mono-temperature can be used for environmental monitoring in our classified area, and that it produces better results in terms of detection of both human strains (sensitive to low temperatures) and environmental strains (sensitive to high temperatures).
This phase also showed that the use of an automated real-time reader facilitates earlier detection of a colony than the human eye, in most cases. (The best time gain was more than 30 hours). However, there are a few exceptions, notably certain fast-growing, fluffy moulds with no specific pigmentation, which the system fails to detect or detects very late. This technology not only ensures reactivity in detection, but also provides the opportunity, by taking snapshots during incubation, to accurately check the count if the box is invaded at the end of incubation.
Future prospects
This study shows that it is possible to choose single-temperature incubation in a pharmaceutical environment, thanks to the efficiency demonstrated in this study. The choice of incubation temperature must be based on data adapted to the specific environment. Site strains are of major interest in this context. To say that 25°C is the only valid temperature is erroneous, but in view of the data generated in our context, it remains the optimum temperature for the detection of contamination in our production environment, thus guaranteeing drug quality and, ultimately, patient safety.
Automation has proved its worth in terms of early colony detection. This point should not be overlooked, as it is crucial for contaminant detection in the classified area to rapidly implement remediation measures.
The future of environmental monitoring lies in the combination of a single incubation temperature and automation.
About the author
Guillaume Pinon
Trained as a microbiologist, Guillaume is currently in charge of the microbiology laboratory for Laboratoires SERVIER Industrie. He has worked for some 20 years for various pharmaceutical manufacturing facilities (sterile injectable and non-sterile powder, solid, liquid formulations) as well as in a laboratory specialised in microbiology for pharmaceutical, cosmetic and biotechnological quality control.
References
- USP <1116> MICROBIOLOGICAL CONTROL AND MONITORING OF ASEPTIC PROCESSING ENVIRONMENTS
- PDA October 2021 Thierry Bonnevay (Sanofi) and Laurent Leblanc (Biomérieux): Suitability of a Single Incubation Temperature for Environmental Monitoring Program
- PDA October 2020: Linda BOELE and Arabela CUIROLO (KYTE): Selection of incubation conditions for environmental monitoring samples at TCF04-Kite pharma EU
Issue
Related topics
Aseptic Processing, Cleanrooms, Drug Manufacturing, Environmental Monitoring, Microbiology
Related organisations
ACM Pharma, Servier, US Food and Drug Administration (FDA), World Health Organization (WHO)