There is more to qPCR than the PCR reaction
Posted: 29 September 2008 | Dr. Jim Huggett, Senior Research Fellow, University College London | No comments yet
The polymerase chain reaction is arguably the most significant technical discovery yet to have been made in the field of molecular biology and genetics, if not all life science. It cannot be overstated how much of an impact this technique has had, resulting in molecular biology becoming an integral part of most biological and medical research fields. PCR is both highly versatile and extremely sensitive, yet with basic training and adherence to key precautions against contamination, it is comparatively simple to perform. However, when conducting PCR, there are a number of additional considerations that are frequently overlooked when investigating the presence or amount of nucleic acid within a sample.
The polymerase chain reaction is arguably the most significant technical discovery yet to have been made in the field of molecular biology and genetics, if not all life science. It cannot be overstated how much of an impact this technique has had, resulting in molecular biology becoming an integral part of most biological and medical research fields. PCR is both highly versatile and extremely sensitive, yet with basic training and adherence to key precautions against contamination, it is comparatively simple to perform. However, when conducting PCR, there are a number of additional considerations that are frequently overlooked when investigating the presence or amount of nucleic acid within a sample.
The polymerase chain reaction is arguably the most significant technical discovery yet to have been made in the field of molecular biology and genetics, if not all life science. It cannot be overstated how much of an impact this technique has had, resulting in molecular biology becoming an integral part of most biological and medical research fields. PCR is both highly versatile and extremely sensitive, yet with basic training and adherence to key precautions against contamination, it is comparatively simple to perform. However, when conducting PCR, there are a number of additional considerations that are frequently overlooked when investigating the presence or amount of nucleic acid within a sample.
While PCR may be easy to get setup in the laboratory, other confounding factors can, if not considered appropriately, make data interpretation very difficult with the potential nightmare of generating completely wrong results. In this series Professor Bustin points out in his article that the PCR step itself is the most robust part of the PCR measurement1. If using quantitative PCR (qPCR) it is possible to measure over eleven orders of magnitude (although one struggles to imagine were this would be necessary) with highly reproducible measurements (coefficience of variation can be less than 10%). But what is frequently overlooked is that there is more to the qPCR measurement than only the qPCR reaction.
This problem is enhanced by the fact that it is the PCR step itself that is frequently the only component that is given any thought. Setting up a new PCR reaction is not a small undertaking as it is fairly complicated to design. Among molecular biologists it is generally accepted that a qPCR reaction must be optimised or at the very least its characteristics (efficiency, detection sensitivity etc.) be included in any publication, it is also becoming frowned upon to simply replicate a previously published PCR assay without any explanation of why this was chosen. However in the literature there is too frequently a complete absence of any other discussion as to the rationale for choice of the various other non-PCR aspects (see Figure 1) required to conduct the PCR.
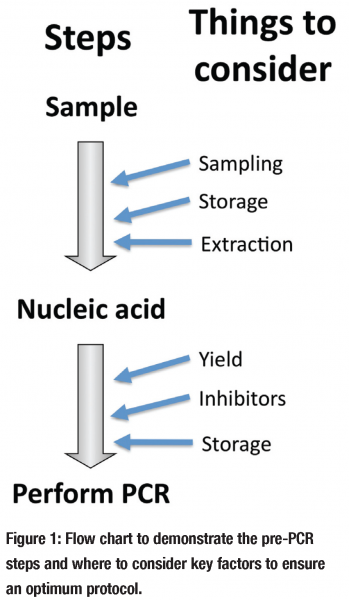

qPCR (and conventional PCR) is dependent on a multistep protocol (see Figure 1) requiring sampling, storage of sample, extraction of sample, the assessment and storage of extract and finally the PCR; additional enzymatic steps are required for reverse transcription qPCR (RT qPCR) and there are a number of other factors that must be considered after the qPCR reaction has been performed, like data normalisation, but there not space to discuss this here and they are covered extensively elsewhere. I will focus on the pre-PCR factors that should be considered when performing qPCR to investigate the presence and quantity of a nucleic acid target within a sample extract. During this article I shall use examples from arguably the two major fields that use qPCR; RNA gene expression analysis by RT qPCR and molecular diagnosis.
Sampling/storage
Sample collection and the means and duration of its storage prior to extraction are seldom discussed in detail and, unless the publication is specifically addressing the subject2, there is usually no data demonstrating that the choice is suitable. Timely fixing of a sample for RNA gene expression analysis is especially important3. This is not only because the RNA is susceptible to degradation by endogenous RNases4 but also because the tissues or cells can remain alive following sampling and may change there expression profile. A good example is when human blood is sampled from a clinic. Practical factors often dictate that there will be a delay between taking the sample and getting it to the laboratory. The various cell types that make up the blood will not necessarily be dead and their expression is likely to change to reflect their new ex vivo status5. There are a number of commercially available reagents that allow the sample RNA profile to be “fixed”. These include PAXgene™ for whole blood and RNAlater® for tissue and cells. These additional reagents can be of great value for certain experimental scenarios, like clinical sampling, but they are expensive. Snap freezing the sample in liquid nitrogen, were possible, is sufficient or frequently the samples can be stored in the lysis buffer that will be used to perform the extraction. Sampling can also lead to the problem of representation; is the sample representative? Biopsy’s may not contain the offending pathogen or tumour, or it may represent such a small percentage of the sample that it is difficult to measure6. The advice for this (as will be apparent from most of this article) is to conduct the preliminary experiments to convince yourself that your chosen strategy is suitable.
It is essential that molecular diagnostic protocols using qPCR to measure both RNA and DNA consider the sampling and storage steps. Where qPCR is used in routine diagnosis, such as in viral identification or load measurements, this must be put in place for the test to be used and forms an integral part of the standard operating procedure (SOP). Unfortunately diagnostic research publications that investigate the use of qPCR as a replacement or compliment to conventional methods, like culture and microscopy, too frequently do not discuss this in detail. Material and methods often provide detailed information about the patient group and rational behind the choice of PCR, with little in between. Good examples are molecular diagnostic studies of tuberculosis (TB) and the tropical diseases where age old microscopic and culture methods represent the techniques of choice and there is little resource available to conduct thorough analyses of newer potential methods. TB is the world’s most prevalent infectious disease infecting 1/3 of the world’s population and killing about 5,000 a day7. Ironically TB is a difficult disease to diagnose. New diagnostic methods could prove highly valuable for the management of this infection and while there are a number of exciting new molecular methods on the market, there is a lack of trust among physicians. I would suspect that this is partly due to TB diagnostic studies that take a single PCR reaction, with no description of evaluation of the pre-PCR, and claim to have evaluated PCR for diagnosis of TB. Without detailed assessment of the sampling, storage and the other steps described below, such a claim cannot be made.
Nucleic acid Extraction
Choice of extraction must be made using other criteria than “it is what we had in the laboratory” or “the company gave us a discount on the kit.” Nucleic acid extraction methods come in a number of guises, yet when discussing this subject with various individuals there is frequently a blind faith that the extraction step must work. Companies like Qiagen offer a huge range of extraction kits for a plethora of different samples and situations. They also provide considerable additional information and expertise as to what to expect from the extract. Despite this, is it worth contacting any extraction provider to establish exactly what they have done to validate their methodologies? Furthermore this may require that the people from the company’s technical support do some investigating themselves, as they may not know the answer. I have on a number of occasions found the asking such questions to be unexpected.
When assessing extractions suitability two things should be considered: yield and removal of inhibitors.
Yield (Amount, Quality and Representation)
What is the amount of nucleic acid being extracted? Is the method of choice the most efficient? How much of the original nucleic acid is recovered and how representative is it? Do any or all of these points actually matter to the specific requirement? There are no single answers to these questions and they very much depend on the specific experimental situation. Nevertheless there are hundreds of extraction methods available and it can considerably improve ones measurement if some time is taken to answer these questions prior to choosing an extraction method.
For molecular diagnosis improving the yield may cause a reduction in diagnostic specificity (false positive) when using conventional PCR if the pathogen is present asymptomatically8; however, using qPCR allows pathogen nucleic acid copy number to be factored into the equation. Consequently, if a disease correlates with load, a “cut off” copy number can be employed below which the individual can be assessed as not having the disease9. How well does the chosen extraction method work with the disease you are interested in? Pathogens can differ from the host in ways that complicate the extraction (e.g. cell walls)10. Were qPCR is used clinically extraction methods are rigorously assessed and form part of a detailed SOP, as with sampling and storage. Extraction controls are usually employed to control for the whole test i.e. if the test fails it maybe due to degradation, poor yield or inhibition. This is discussed in more detail below.
Sample quality is an area that has been very heavily pressed in the field of RNA gene expression analysis and it is now common place for RNA quality to form part of the methods of a paper using RT qPCR. This is most frequently performed by gel electrophoresis showing two clear ribosomal bands when the sample quality is good. As the RNA degrades these bands tend to become more diffuse with a smear at the bottom of gel when degraded. The rRNA bands are ideal for visualisation, as they not only form ~80 % of the RNA fraction, but those that can be easily seen, comprise of two distinct sizes that appear as distinct bands. A potential problem occurs, however, as it is the protein coding messenger RNA (mRNA) and not the rRNA fraction that is usually measured by RT qPCR and degradation of rRNA does not necessarily mean degradation of the mRNA11. While this represents a good control step this is largely due to the absence of better alternatives. Recently, an idea proposed by Nolan et al. uses a 3’ 5’ assay to measure the mRNA degradation12 of the specific RNA(s) under investigation. This is a much more favourable approach, but is more complicated requiring at least two assays for each RNA measured. This approach can also be confused by the presence of inhibitors, which should also be controlled for, at least during assay setup.
Inhibition
If you conduct qPCR and answer yes to the following question: do you specifically assess your extracts for inhibition? Then you represent a small minority. Molecular biologists know that the PCR reaction is susceptible to inhibition yet few will employ specific experiments to assess this. What is often conducted is the use of extraction controls that report a problem when they are not detected. Extraction controls do not stipulate what is the cause of the problem, but this information is often not needed. Clinical diagnosis uses this approach as such controls are central to making any clinical diagnosis and inhibition is a factor considered by this. These control reactions comprise either different PCR reactions or modified versions of the same reaction and are co-extracted with the sample of interest thus controlling for nucleic acid recovery and removal of inhibitors.
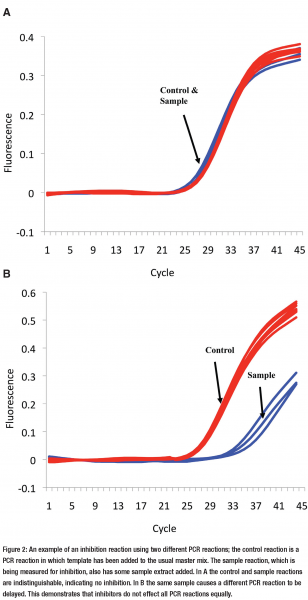
Diagnostic research has suffered here as well. There are many studies that have taken a qPCR reaction and looked at a set of patient samples, without any question of yield or inhibition, found they do not work, and concluded that PCR is not suitable for the disease in question. One simply cannot state that qPCR is not suitable for diagnosing a particular disease unless inhibition (and the other points outlined in Figure 1) is considered because the negative result may be false. A simple way to assess inhibition is to perform an PCR in which you add a known quantity of template to the PCR reaction master mix then compare your extract to a water control (see Figures 2a & 2b on page 18). This method, termed inhibition reaction, is artificial and is only establishing the effect of the extract on the artificial assay’s performance.

The notion that, and method by which, inhibitors can be analysed has recently been encouraged for RT qPCR13. However many will argue that as gene expression studies usually included the measurement of a reference gene (which may or may not be a house keeping gene) for normalisation, this will also control for inhibition, in much the same way as the extraction controls are used in diagnosis. While this assumption may seem fair, it is becoming increasingly clear that it may be an assumption too far.
Using extraction controls or inhibition reactions to measure inhibition assumes one fundamental thing:
If I use reaction A as an inhibition reaction (or extraction control) any reported inhibition will also inhibit reaction B to the same extent.
Assuming that the inhibitors have an equally effect all reactions underlies all situations where more than one reaction is performed on a sample and those reactions are then compared. This applies when performing specific inhibition experiments or when using extraction controls, reference genes or any situation were two PCR reactions are measured within a sample and compared.
But this assumption that all reactions are equally effected by inhibitors is wrong14. It is possible for one reaction to be completely unaffected by the inhibitors, within a sample extract, while another is severely inhibited (see Figures 2a & 2b). However as extraction methods are employed to remove inhibitors, how relevant is this observation to the every day laboratory? Our laboratory is beginning to ask this very question and we are finding that many commercial and in house extraction methods frequently co-purify inhibitors. Inhibition can have a subtle effect altering the result by a little or a major effect (see Figure 3). This is likely to vary depending on the PCR reaction and sample extract.
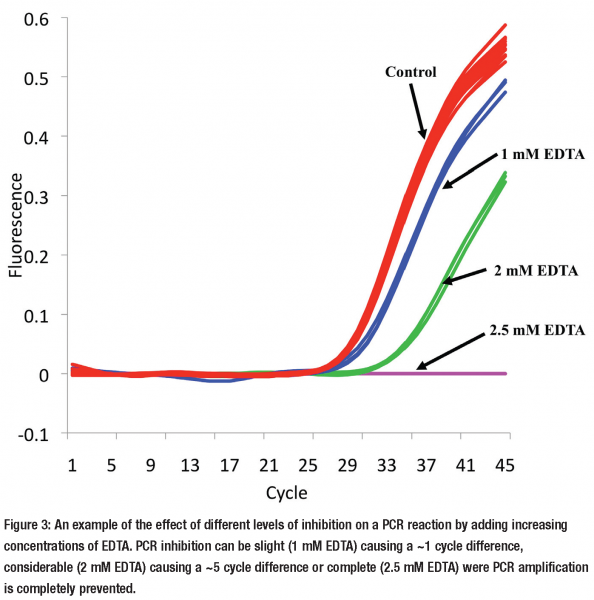

In our laboratory we routinely assess for PCR inhibition using inhibition reactions. However, this requires the development of another assay and confirmation that it is equally affected by inhibitors as the reaction(s) of interest14. A simpler method we also employ is to dilute the sample. When a sample gives a result of 5,000 and you dilute it 10 fold and repeat the experiment, if the new result is > 500 there is an inhibitor present. This touches on another point; if you have a number of extracts that have inhibitors present, one of the ways of getting around this is to dilute the sample. This dilutes the inhibitor and can remove the inhibitory effect. There is of course a trade off where by you eventually dilute the DNA of interest so much that it is not longer measurable. If this happens repeating the sampling with a more appropriate extraction method is the only solution.
Storage of nucleic acid
How to store nucleic acids may seem an odd inclusion in this section as it is fairly well established that well purified nuclease free nucleic acid extracts are generally very stable. In our laboratory we store nucleic acids at -20 °C for most situations, with < -70 °C for long term storage (> 1 month). Qiagen recommends RNA storage in RNase free water at -20 °C or -70 ° reporting no degradation after a year. They recommend that if DNA storage is in DNase free water the pH should be > 7.0, favouring 10 mM Tris-Cl. 0.5 mM EDTA, pH 9.0. This is a variation on the commonly used 10mM Tris-HCI, 1 mM EDTA, pH 7.4-8 (TE) buffer, which is also suitable. Using buffers that contain EDTA protect nucleic acids from Mg 2+ dependent degradation, like DNase I, but the inclusion of EDTA can cause problems with subsequent enzymatic experiments like the PCR. Nucleic acids are not stable after repeat freeze thawing15, so while sample aliquoting is laborious, it is essential to ensure that your samples can be measured and then re-assessed without the risks of freeze thaw damage.
Conclusion
The literature suggests there is not enough attention given to the pre-PCR steps when conducting qPCR. Either this is not considered or not included within the material and methods. The scientists, who conduct the research, cannot take all the blame as space constraints, pressed by journal editors, limits how much information can be submitted. With the introduction of open access and increasing options of submitting additional accompanying information online, this problem will hopefully be resolved.
Considering how one obtains and stores a sample and then extracts the nucleic acids is essential. Furthermore quality assessment of the nucleic acid can benefit the final measurements and it is recommended that some assessment of inhibitors is performed to rule out or manage their presence. As stated the qPCR method is highly versatile, however if the pre-PCR steps are not considered then the full potential of this methodology is at danger of not being realised.
Acknowledgements
I am grateful to Nina Witt and Tanya Novak for critical review of this manuscript.
References
- Bustin SA. The real-time reverse transcription polymerase chain reaction – treat with caution European Pharmaceutical Review 2007.
- Grant PR, Kitchen A, Barbara JA, Hewitt P, Sims CM, Garson JA, et al. Effects of handling and storage of blood on the stability of hepatitis C virus RNA: implications for NAT testing in transfusion practice. Vox Sang 2000;78(3):137-42.
- Madejon A, Manzano ML, Arocena C, Castillo I, Carreno V. Effects of delayed freezing of liver biopsies on the detection of hepatitis C virus RNA strands. J Hepatol 2000;32(6):1019-25.
- Bustin SA, Nolan T. Template Handling, Preparation and Quantification. In: Bustin SA, ed. A-Z of Quantitative PCR. La Jolla: International University Lie; 2004.
- Tanner MA, Berk LS, Felten DL, Blidy AD, Bit SL, Ruff DW. Substantial changes in gene expression level due to the storage temperature and storage duration of human whole blood. Clin Lab Haematol 2002;24(6):337-41.
- Berthon P, Dimitrov T, Stower M, Cussenot O, Maitland NJ. A microdissection approach to detect molecular markers during progression of prostate cancer. Br J Cancer 1995;72(4):946-51.
- Zumla A. Tuberculosis–the tide can be turned, the battle can be won. J R Soc Med 2008;101(3):100-1.
- Huggett JF, Miller RF, Taylor MS, Costello AM, Zumla A. Problems of developing molecular diagnostic tests for opportunistic pathogens: the example of Pneumocystis jirovecii. J Eukaryot Microbiol 2006;53 Suppl 1:S85-6.
- Huggett JF, Taylor MS, Kocjan G, Evans HE, Morris-Jones S, Gant V, et al. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax 2007;63(2):154-9.
- Dietrich G, Schaible UE, Diehl KD, Mollenkopf H, Wiek S, Hess J, et al. Isolation of RNA from mycobacteria grown under in vitro and in vivo conditions. FEMS Microbiol Lett 2000;186(2):177-80.
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 2006;27(2-3):126-39.
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc 2006;1(3):1559-82.
- Nolan T, Hands RE, Ogunkolade W, Bustin SA. SPUD: A quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem 2006;351(2):308-10.
- Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, et al. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes 2008;1(1):70.
- Moriya Y, Nakamura T, Okamura N, Sakaeda T, Horinouchi M, Tamura T, et al. Comparison of synthetic DNA templates with authentic cDNA templates in terms of quantification by real-time quantitative reverse transcription polymerase chain reaction. Biol Pharm Bull 2006;29(3):535-8.





