Chromatin: cracking the chemical code
Posted: 2 August 2008 | Colin Reynolds, School of Biomolecular Sciences, Liverpool John Moores University; James Nicholson and John Baldwin, Biology & Medicine College, STFC Daresbury Laboratory | No comments yet
Chromatin is a nucleoprotein complex, found in eukaryotic organisms, consisting of approximately 50% DNA and 50% histone proteins. The histones undergo a number of reversible covalent chemical modifications (for example, acetylation, methylation, phosphorylation), especially in the N-terminal regions. These chemical changes prepare chromatin for a range of essential cellular processes and understanding the code for the many possible combinations of modifications, in terms of their function, is currently an area of intense research and is driving developments in molecular cancer therapies. Several molecules that modulate chromatin modifications have already been developed as commercially available cancer treatments, while many others are undergoing clinical trials.
Chromatin is a nucleoprotein complex, found in eukaryotic organisms, consisting of approximately 50% DNA and 50% histone proteins. The histones undergo a number of reversible covalent chemical modifications (for example, acetylation, methylation, phosphorylation), especially in the N-terminal regions. These chemical changes prepare chromatin for a range of essential cellular processes and understanding the code for the many possible combinations of modifications, in terms of their function, is currently an area of intense research and is driving developments in molecular cancer therapies. Several molecules that modulate chromatin modifications have already been developed as commercially available cancer treatments, while many others are undergoing clinical trials.
Chromatin is a nucleoprotein complex, found in eukaryotic organisms, consisting of approximately 50% DNA and 50% histone proteins. The histones undergo a number of reversible covalent chemical modifications (for example, acetylation, methylation, phosphorylation), especially in the N-terminal regions. These chemical changes prepare chromatin for a range of essential cellular processes and understanding the code for the many possible combinations of modifications, in terms of their function, is currently an area of intense research and is driving developments in molecular cancer therapies. Several molecules that modulate chromatin modifications have already been developed as commercially available cancer treatments, while many others are undergoing clinical trials.
Genomic DNA is packaged in the limited space of the nuclei of eukaryotic cells in the form of chromatin, a DNA-histone complex. The histones are small, positively charged proteins, which interact with DNA to form nucleosomes, the repeat unit of chromatin1. The core of the nucleosome structure, or nucleosome core particle (NCP), as well as the isolated histone octamer have been determined by X-ray crystallography2,3 and have shown to consist of 146 base pairs of DNA wrapped around the histone octamer, containing two copies each of the highly conserved core histones; H2A, H2B, H3 and H4. In the full nucleosome, a fifth, more variable, histone, H1 (H5 in birds), known as linker histone, interacts with the DNA at the entry and exit points from the nucleosome. There is also a variable species-dependent 0 to 60 base-pair section of linker DNA, which connects one nucleosome to the next in a zigzag fashion. Each core histone contains a C-terminal globular domain as well as an N-terminal tail rich in arginine and lysine residues and these are the main sites of chemical or post-translational modifications (PTMs) shown in Figure 1. H2A is unique amongst the core histones in that it also contains a C-terminal tail. In recent years, the application of high-resolution mass spectrometry has allowed the detailed mapping of the PTMs of the histone proteins4. Biochemical kits are now available that use the chromatin immuno-precipitation (ChIP) method to relate modified amino-acid residues in a histone to the DNA sequence in the corresponding nucleosome. The histone tails are mobile and are on the surface of the nucleosome. They play an important role in the formation of higher-order chromatin structures and are involved in the mechanism for condensing and decondensing chromatin, which is essential for controlling gene expression and cell function.
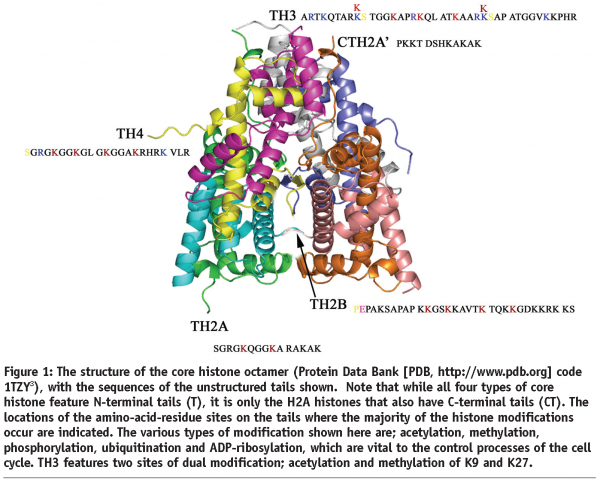


A change in gene-expression profile that occurs without any alteration in the genomic DNA sequence of a cell is an example of epigenetics. The altered pattern of gene expression is important to cellular differentiation and development and is inherited by daughter cells, thereby maintaining the integrity, specifications, and functions of a given cell type. The genome of eukaryotic cells carries epigenetic information that is mainly encoded by methylation of cytosines of DNA and it is becoming increasingly clear that covalent PTMs of histones play a significant part5, thus allowing regulation of genome accessibility for the various DNA-related processes such as recombination, repair, replication and transcription. The conversion of these patterns of histone modifications (the ‘histone code’)6 into relevant biological signals is achieved by a variety of histone-interacting proteins and chromatin-modifying machines, which contain specific binding domains such as the bromodomain and chromodomain (see Figures 3 and 4). Here we consider the different types of PTMs of histones and their implications for chromatin structure and function, as well as their potential wider applications in fighting diseases such as cancer.


Acetylation
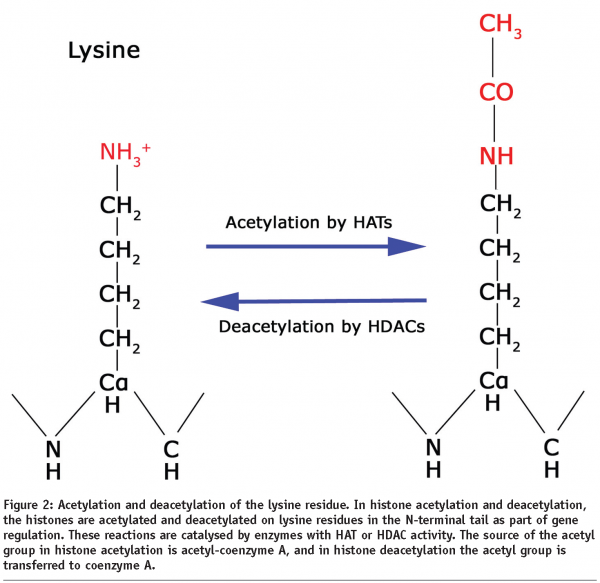
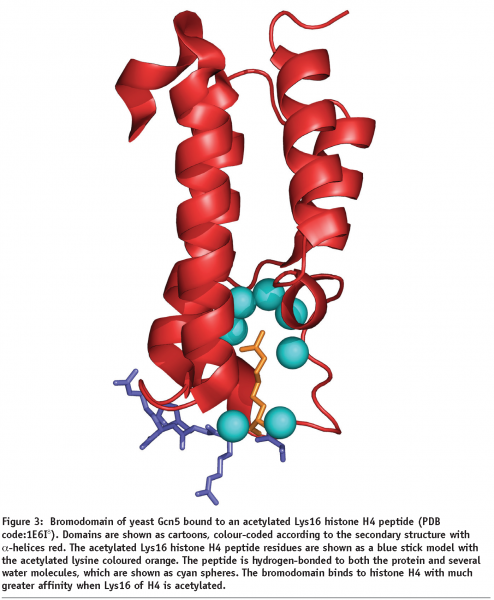
Acetylation is a widely occurring PTM found in all animal and plant species. Lysines at the N-termini of the histones may be modified by enzymes known as histone acetyl transferases (HATs), which replace a hydrogen atom in the amino group of the lysine side chain by an acetyl group from acetyl-coenzyme A, thus removing its positive charge and reducing its affinity for DNA binding (Figure 2). For example, the TAFII250 subunit of the TFIID transcription factor, which is important for basal transcription of many genes, has been shown to have HAT activity7. This modification occurs particularly for histones H3 (at lysines 9, 14, 18 and 23) and H4 (at lysines 5, 8, 12 and 16), where hyperacetylated forms containing several acetyl groups have been shown to be localised preferentially in active genes whereas under-acetylation (hypoacetylation) is characteristic of transcriptionally inactive genes. Different patterns of acetylation may be necessary for optimal activation of different sets of genes. Several activating factors that bind to acetylated lysine regions of histone proteins contain a region known as the bromodomain that binds to histones with much greater affinity when specific lysines in the histones are acetylated (Figure 3)8. Binding of these bromodomains to acetylated histones results in a more open chromatin structure and transcriptional activation. Enzymes which deacetylate histones have also been characterised and are known as histone deacetylases or HDACs. For example, the nuclear receptor co-repressor, which is a bromodomain-containing activating factor that mediates the inhibiting effect of the thyroid hormone receptor on transcription when thyroid hormone is absent, has been shown to associate with a protein complex having HDAC activity9. Thus, activating factors can direct the acetylation of histones, thereby opening up (decondensing) the chromatin, whereas inhibiting factors can give rise to histone deacetylation, resulting in a more tightly-packed (condensed) chromatin structure.
Errors in the function of HDACs can lead to the proliferation of cancerous cells and, therefore HDAC inhibitors (HDACIs) have been targeted by academic and commercial R&D groups as potential cancer therapies.
Methylation
Histones can be modified by addition of a methyl group to arginines and to lysines by specific enzymes (histone methyl transferases, HMTs), using S-adenosyl methionine as the methyl donor. For example, residue Lys9 of H3, is a substrate for a specific histone methyl transferase enzyme – this particular modification is associated with transcriptionally inactive DNA10. In contrast, methylation of Lys79 on the globular region of histone H3 has been shown to have the potential role to mark open or transcriptionally active chromatin (euchromatin) regions, possibly by changing the association between nucleosomes, or by preventing the binding of protein factors that are necessary for chromatin silencing10. Another enzyme, known as coactivator-associated arginine methyltransferase 1 (CARM1), methylates histone H3 at Arg17 and Arg26 upon hormonal activation of nuclear receptors11. CARM1 activity towards histone H3 Arg17 is potentiated by pre-acetylation at Lys18, suggesting a possible ‘cross-talk’ mechanism between histone acetylation and methylation, whereby one type of histone modification can modulate another form of modification.
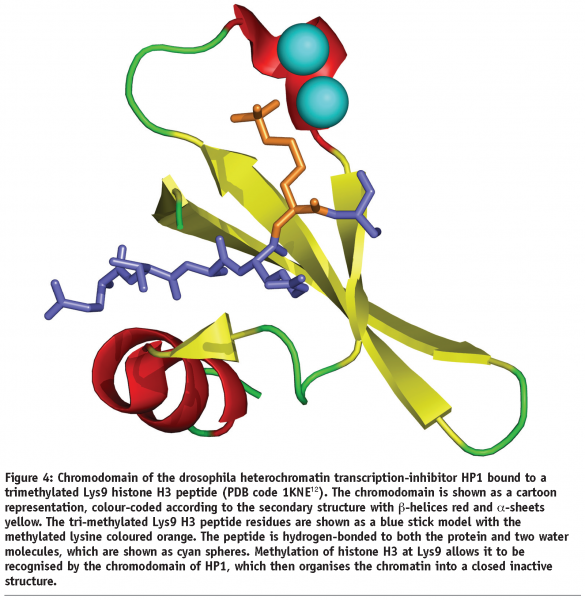
Chromodomains (Figure 4) are found in many proteins that inhibit transcription, such as heterochromatin protein 1 (HP1), which binds with methylated amino acid residues on histone H312. For example, methylation of histone H3 on Lys9 allows it to be recognised by the chromodomain of HP1, which then organises the chromatin into a closed inactive structure.
Methylation was for many years believed to be an irreversible and a stable modification of histones. However, in the last few years several enzymes have been discovered to demethylate mono- and di-methylated lysine residues of histone H3 as well as monomethylated arginines via either amine oxidation or deamination, respectively. For example the JmjC domain-containing histone demethylase 1 (JHDM1), which is conserved from yeast to human, has been demonstrated to demethylate mono- and di- but not tri-methylated H3 K36 via hydroxylation of the methyl moiety within the methylated lysine residue13.
DNA methylation within the nucleosome is the primary epigenetic modification, for example, one that can be inherited without changing the DNA sequence, which can result in gene silencing and loss of function. DNA methylation involves the addition of a methyl group directly onto DNA, for example, to carbon 5 of the cytosine pyrimidine ring.
Citrullination
Citrullination or deimination is the term used for the post-translational modification of arginine into citrulline (Figure 5). This reaction is performed by enzymes called peptidylarginine deiminases (PADs). The conversion of arginine into citrulline can have important consequences for the structure and function of proteins, since arginine is positively charged at a neutral pH, whereas citrulline is uncharged. This increases the hydrophobicity of the protein, leading to protein unfolding.
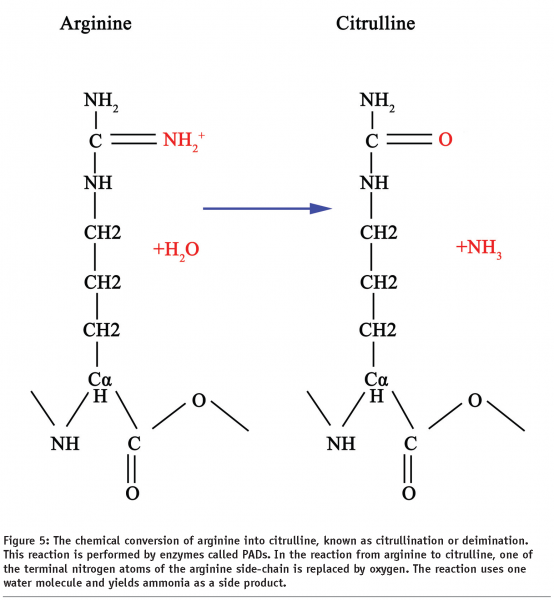

Some forms of PAD have been proposed to effectively repress transcription by reversing the possible epigenetic histone modifications mediated by arginine methyltransferases. Hence PADs operate effectively as histone demethylases.
For example, the methyltransferase PRMT1 methylates histone H4 at Arg3, leading to chromatin remodelling and increased binding to the transcriptional complexes, resulting in increased transcription14. If PADI4 is recruited during this process it catalyses the conversion of methylarginine into citrulline releasing methylamine. Citrullination of histone methylarginine residues decreases the ability of transcriptional machinery to bind to chromatin, thereby repressing transcription.
Phosphorylation
Phosphorylation is one of the most important processes in biology. Many enzymes and receptors are regulated by phosphorylation and dephosphorylation. Phosphorylation is catalysed by a family of enzymes known as protein kinases, whereas phosphatases are responsible for dephosphorylation. In chromatin, protein kinases phosphorylate the –OH group of histone serines and threonines giving an O-phosphate linkage, whereas protein phosphatases can reverse this process. The phosphate donor is usually ATP, GTP or cyclic AMP.
The breast cancer protein BRCA1 contains BRCT domains, either singly or in tandem, which are associated with DNA damage as well as cell-cycle checkpoints and have been shown to bind the phosphorylated histone variant H2A.X15. Following DNA damage, the serine residue 139 of H2A.X is phosphorylated, resulting in the recruitment of many DNA-damage signaling and repair proteins. Structural studies have been undertaken, which have yielded clues to the molecular events that couple DNA damage to chromatin signaling via phosphorylation of the histone proteins.
Experiments have shown that when cells are exposed to heat shock, phosphorylated histone H3 is concentrated at the heat-shock gene loci which are being actively transcribed, but is depleted from other loci which are silenced following heat shock16. Other experiments have shown that when cells are stimulated with growth factors, histone H3 is phosphorylated on Ser10 and the modified histones are localised to genes which become transcriptionally active16. Interestingly, such growth factor-induced phosphorylation of H3 does not occur in human patients with Coffin-Lowry syndrome (gene mutation resulting in severe mental impairment) who lack the phosphorylation enzyme17. It has been shown that phosphorylation of Ser10 in H3 enhances the ability of HATs to acetylate Lys14 of H3 and HMTs to methylate Lys4, thus organising the chromatin into an accessible form, favourable for transcription18. This is a further example of chromatin ‘cross-talk’ for example, different combinations of chemical histone modifications acting synergistically in chromatin function19. Some molecular cancer therapies are targeted at the tyrosine kinases that mediate protein phosphorylation, such as the commercially available tyrosine kinase inhibitor Herceptin.
Ubiquitination and SUMOylation
Ubiquitin is a small regulatory protein (ubiquitous in eukaryotes) that is routinely used by cells to label old, damaged or misfolded proteins for destruction in a proteasome. Ubiquitination (sometimes called ubiquitylation) refers to the covalent attachment of ubiquitin to another protein molecule. Ubiquitin can form a covalent isopeptide linkage with histones H2A or H2B via its C-terminal carboxyl group, which joins the amino group of lysine residues20. This modification (to Lys119 of H2A or Lys120 of H2B) reduces the net positive charge on the histone. Only approximately 10% of H2A in a cell is ubiquitinated and this modification is associated with repression of gene expression. In contrast, ubiquitination of H2B (about 2% of total) stimulates gene expression since it promotes the methylation of H3 on Lys4 and Lys79, resulting in an open chromatin structure. This is a more sophisticated example of ‘cross-talk’, where one form of modification is modulating another type of modification on a completely different type of histone protein.
Lysine residues on histones can also be modified by addition of SUMO (Small Ubiquitin-related Modifier), resulting in transcriptional repression via deacetylation of histones giving a more closed chromatin structure.
SUMOylation is a post-translational modification involved in several cellular processes as well as transcriptional regulation, including apoptosis, protein stability, stress response and cell-cycle progression. The SUMO proteins are similar in structure to ubiquitin, and SUMOylation is controlled by enzymatic processes analogous to those of ubiquitination. However, in contrast to ubiquitin, SUMO is not used to label proteins for degradation.
ADP-ribosylation
ADP-ribosylation is a modification that involves the attachment of ADP-ribose to a protein. The activities of the enzymes that control this process are dependent upon DNA strand breaks and ADP-ribosylated histones have been observed as an intermediate structure in nuclear processes involving DNA strand breaks. Therefore it is believed that ADP-ribosylation of core histones (predominantly H2B) plays an important role in DNA repair21. ADP-ribosylation of core histones can result in localised unfolding of the chromatin higher-order fibre structure. It is thought that the binding of a polymer form of ADP-ribose to histone H2B could aid the dissociation of H3 and H4 from DNA during the repair process.
Polymeric ADP-ribose bound to histones may also act as a type of chaperone molecule during nucleosome formation, protecting the charged N-terminal tails while the hydrophobic globular domains of the histones associate and organise the DNA into a nucleosome structure.
Histone variants
Several variant forms of specific histones (excluding H4) are encoded by distinct genes, for example, there are three H3 variants in mammals, H3.1, H3.2 and H3.3, whereas H2A has at least three variants; H2A.Z, H2A.X and macroH2A22. Both H2A.Z and H3.3 have been shown to be preferentially localised in the active genes of euchromatin, yet potentially involved in the formation of the genetically inactive heterochromatin. H2A.X binds to DNA with double strand breaks and marks regions undergoing DNA repair. MacroH2A, which has a very long C-terminal tail, has been shown to associate specifically with the inactive, transcriptionally silent and heterochromatic X chromosome in female mammals. The histone variant H3-like CENP-A is essential for chromosomal centromere structure and function.
The histone variants feature many non-conserved amino acid replacements at the sites of post-translational modification in both the tails and globular domains. This leads to a radical change in the pattern of chemical interactions for these variants compared with the ‘native’ histones, resulting in the range of alternative functions described above.
However, the variants are only required to exist in tiny proportions compared with the usual core histones in order to fulfil their niche functions.
Concluding remarks
The many chemical modifications associated with possible epigenetic expression that occur within chromatin are fundamental in understanding gene regulation, cellular processes and in particular errors in cellular events. Indeed, some cancers have been linked to chemical modifications of histones and DNA. For example, malfunctions in the HDACs may result in these enzymes proliferating and therefore disproportionately down-regulating the genes that control the normal processes of programmed cell death, known as apoptosis. If apoptosis is inhibited in cancerous cells they will multiply and grow into a tumour. Consequently HDAC inhibitors (HDACIs) such as SAHA, trichostatin A and apicidin have been vigorously targeted as potential molecular therapies for cancer23.
Chemical processes that remove methyl groups (demethylases) from DNA and histones are also being targeted for cancer therapies, since DNA methylation and histone deacetylation have been shown to act synergistically to promote the onset of some tumours, while histone methylation can be a crucial factor in regulating DNA methylation24,25.
Understanding the myriad of interacting chemical modifications in chromatin is essential for both understanding the processes of genetic inheritance and expression, and fighting diseases like cancer at the molecular level. Current research is focused on cracking the chemical code of chromatin; both the functional patterns of modifications and their cross-talking effects, in order to allow the development of novel therapeutic compounds.
Acknowledgments
The authors thank all their colleagues and students (past and present) within the chromatin group of Liverpool John Moores University and STFC Daresbury Laboratory for all their enthusiasm and hard work over the years. Particular thanks go to Stan Lambert and Chris Wood.
References
- J. M. Nicholson and C.M.Wood, (Editors), Chromatin Structure and Function. ISBN: 81-7895-234-3. Transworld Research Network, 2006.
- K. Luger et al, Structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, pp. 251-260, 1997.
- C.M. Wood, et al High-resolution (1.9 Å) structure of the native histone octamer. Acta Cryst. F6, pp. 541-545, 2005.
- A. Villar-Garea and A. Imhof, The analysis of histone modifications. Biochimica et Biophysica Acta 1764, pp. 1932-1939, 2006
- K.P. Nightingale et al, Histone modifications: signaling receptors and potential elements of a heritable epigenetic code. Current Opinion in Genetics & Development 16, pp. 125-136, 2006.
- B.M. Turner, Cellular memory and the histone code. Cell 111, pp. 285-291, 2002.
- R.T. Utley et al, Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394, pp. 498-502, 1998.
- D.J. Owen et al, The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO Journal 19, pp. 6141-6149, 2000.
- J.J. Yu et al, A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation, EMBO Journal 22, pp. 3403-3410, 2003.
- O.J. Rando, Global Patterns of histone modifications. Current Opinion in Genetics & Development 17, pp. 1-6, 2207.
- D.H. Cheng et al, The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Molecular Cell 25, pp. 71-83, 2007.
- S.A. Jacobs, and S. Khorasanizadeh, Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, pp. 2080-2083, 2002.
- J. Schneider and A. Shilatifard, Histone demethylation by hydroxylation: chemistry in action. ACS Chemical Biology 1, pp. 75-81, 2006.
- S.M. Huang et al, Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes & Development 19, pp. 1885-1893, 2005.
- J.C. Morales et al, Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. Journal of Biological Chemistry 278, pp. 14971-14977, 2003.
- S.J. Nowak et al, Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Molecular and Cellular Biology 23, pp. 6129-6138, 2003.
- Ausio, J., Levin, D.B., de Amorim, G.V., Bakker, S. and Macleod, P.M. Syndromes of disordered chromatin remodeling. Clinical Genetics 64 (2003), 83-95.
- A. Clements et al, Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Molecular Cell 12, pp. 461-473 2003.
- W. Fischle et al, Histone and chromatin cross-talk. Current Opinion in Cell Biology 15 (2003), pp. 172-183, 2003.
- M.A. Osley, H2B ubiquitylation: the end is in sight. Biochimica et Biophysica Acta-Gene Structure and Expression 1677, pp.74-78, 2004.
- L. Virag and C. Szabo, The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacological Reviews 54, pp. 375-429, 2002.
- J.M. Nicholson et al, Structural molecular biology of the histone octamer. Recent Research Developments in Molecular Biology 1, pp. 165-187. ISBN 81-271-0032-3, 2003.
- W.S. Xu, Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 26, pp. 5541-5552, 2007.
- W. An, Histone acetylation and methylation: combinatorial players for transcriptional regulation. Subcellular Biochemistry 41, pp. 351-69, 2007.
- M. Esteller, Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Reviews Genetics 8, pp.286-98, 2007.




