DNA sequencing technologies and emerging applications in drug discovery
Posted: 13 December 2011 |
In recent years, the development of Next Generation DNA Sequencing (NGS) technology has significantly impacted molecular biology research, resulting in many new insights and discoveries. NGS technology goes beyond traditional DNA sequencing with applications that reach across the central dogma of molecular biology from DNA to RNA and protein science. Drug discovery is beginning to benefit from the diversity of NGS, with applications in evidence across various therapeutic areas, such as oncology, immunology and infectious diseases.
DNA is the molecule of life, containing the information for the synthesis of RNA molecules and proteins, which in turn form structural components of the cell or catalyse essential biochemical processes. Understanding the sequence of DNA, which is made from the four basic building blocks or ‘nucleotides’, A,G,C and T, has resulted in great insights and discoveries in cellular biology, pathology and disease, culminating in the human genome project, which achieved the remarkable feat of determining the sequence of the three billion bases of the human genome.
The field of DNA sequencing has witnessed some key milestones in technology develop – ment since the description of the first revolutionary DNA sequencing techniques in 19771,2. The Sanger dideoxy sequencing method, discovered by the Nobel Laureate Fred Sanger, underwent the most significant improvements and became the first automated sequencing platform in the late 20th century. Advancements in the Sanger process were partly motivated by the advent of the USD 3 billion Human Genome Project, which required the development of high-throughput tech – niques3,4 (Figure 1A).
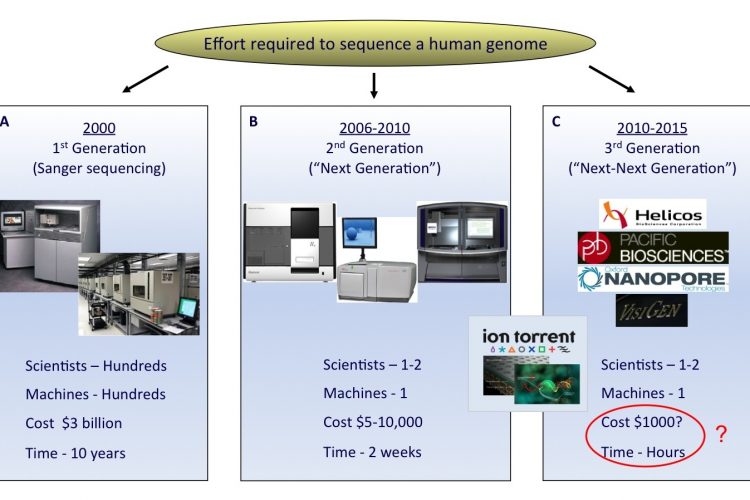

FIGURE 1 The rapid evolution of sequencing technologies. A. First generation Sanger sequencing technology. B. Second ‘Next’ generation massively parallel sequencing technology (454 Sequencing © Roche Diagnostics) C. Third ‘Next-Next’ generation single molecule, real-time sequencing technology. In the coming years, second or third generation technologies may develop to an extent where a human genome can be sequenced for a USD 1,000 in a matter of hours
In recent years, the development of Next Generation DNA Sequencing (NGS) technology has significantly impacted molecular biology research, resulting in many new insights and discoveries. NGS technology goes beyond traditional DNA sequencing with applications that reach across the central dogma of molecular biology from DNA to RNA and protein science. Drug discovery is beginning to benefit from the diversity of NGS, with applications in evidence across various therapeutic areas, such as oncology, immunology and infectious diseases.
DNA is the molecule of life, containing the information for the synthesis of RNA molecules and proteins, which in turn form structural components of the cell or catalyse essential biochemical processes. Understanding the sequence of DNA, which is made from the four basic building blocks or ‘nucleotides’, A,G,C and T, has resulted in great insights and discoveries in cellular biology, pathology and disease, culminating in the human genome project, which achieved the remarkable feat of determining the sequence of the three billion bases of the human genome.
The field of DNA sequencing has witnessed some key milestones in technology develop – ment since the description of the first revolutionary DNA sequencing techniques in 19771,2. The Sanger dideoxy sequencing method, discovered by the Nobel Laureate Fred Sanger, underwent the most significant improvements and became the first automated sequencing platform in the late 20th century. Advancements in the Sanger process were partly motivated by the advent of the USD 3 billion Human Genome Project, which required the development of high-throughput tech – niques3,4 (Figure 1A).
Despite automation, there still remained a throughput limitation of the Sanger technique when conducting large scale sequencing projects. Recent significant advances have addressed this, resulting in the development of next-generation sequencing technology (Figure 1B).
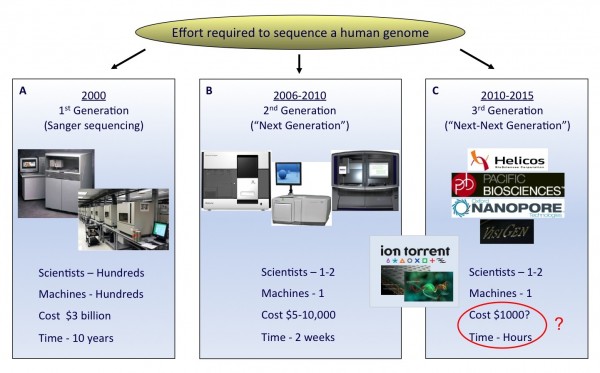

FIGURE 1 The rapid evolution of sequencing technologies. A. First generation Sanger sequencing technology. B. Second ‘Next’ generation massively parallel sequencing technology (454 Sequencing © Roche Diagnostics) C. Third ‘Next-Next’ generation single molecule, real-time sequencing technology. In the coming years, second or third generation technologies may develop to an extent where a human genome can be sequenced for a USD 1,000 in a matter of hours
Next Generation Sequencing Technologies
The first NGS technology to be developed was based on the novel pyrosequencing method5 and was commercially released as the 454 sequencing platform in 20056,7. Additional platforms followed including the Solexa/ Illumina and SOLiD/Life Technologies sequencers (Figure 1B). Although differing in their chemistries and processes, the platforms have broadly similar workflows. The NGS method begins by shearing genomic or cDNA molecules into small fragments, followed by massively parallel PCR amplification and sequencing of individual DNA molecules to produce short read DNA sequences8. These short reads are then aligned by informatics methods which look for overlaps between reads to reconstruct the sequence of the starting template DNA molecule. The 454 sequencing method employs sequential enzymatic incorporation of nucleotides (Figure 2A), where incorporation releases inorganic pyrophosphate, which is subsequently converted into a chemi – luminescent signal9. This signal is detected by a charge-coupled device (CCD) camera and converted into a DNA sequence, in which the light intensity is proportional to the number of incorporated nucleotides10. In contrast, the Illumina technology is based on an alternative sequencing-by-synthesis approach, in which nucleotides have fluorescently labelled reversible terminators attached11 (Figure 2B). The reversible terminators are sequentially incorporated into the growing DNA strand, and imaged to identify the incorporated base. The terminator moiety is then removed to allow for the incorporation of the next reversible terminator. In determining which NGS tech – nology to use, important factors include cost per run, sample preparation complexity, run time, simplicity of data analysis and read lengths generated13.
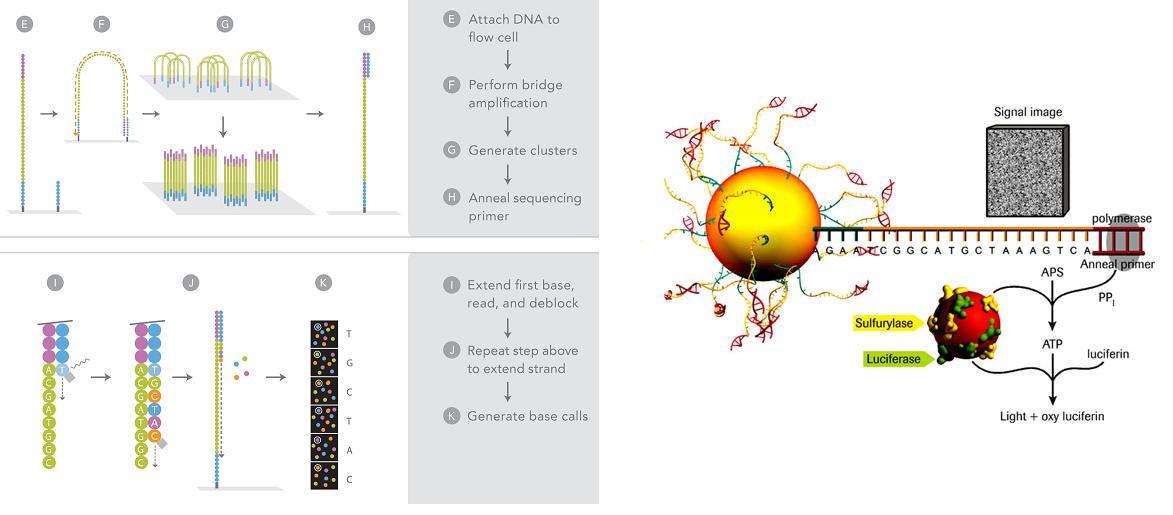

FIGURE 2 Next Generation Sequencing Technology Platforms. Each NGS platform is able to perform massively parallel clonal PCR amplification followed by DNA sequencing. (A) In 454 sequencing, nucleotide incorporation is detected by a light emitting luciferase reaction called pyrosequencing. (© Roche Diagnostics) (B) The Illumina sequencing method uses solid phase amplification of DNA molecules followed by incorporation of fluorescently labelled nucleotides (© Illumina)
The arrival of third generation sequencing technology, in which single DNA molecules are sequenced in real time, promises even faster sequencing with higher data outputs per machine run12 (Figure 1C). These platforms, which include nanopore sequencing and technologies which monitor polymerase base incorporation in real-time, could provide additional benefits, including longer read lengths, rapid run times and reduction in the amount of starting sample required.
Applications in drug discovery
The application of NGS technology within academic laboratories has been rapid and has resulted in many new exciting discoveries. Here we describe some current and potential applications of NGS technology in the drug discovery and development process (Figure 3).
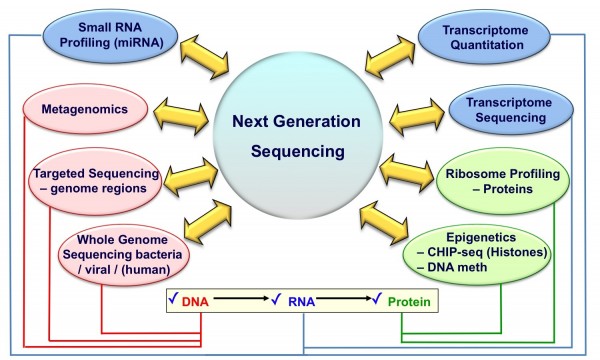

FIGURE 3 Applications of Next Generation Sequencing Technologies. Listed are some of the different applications possible on a single NGS sequencing machine which span across the central dogma of molecular biology (‘DNA makes RNA makes protein’)
Whole genome DNA sequencing
One of the areas where NGS has had a large impact is in sequencing of whole genomes. Whole genome sequencing using NGS allows great depth of sequence coverage in one machine run, which substantially reduces both time and cost as compared to traditional Sanger sequencing.
Such is the throughput of the technology that whole bacterial and viral genomes can now be routinely sequenced in one experiment, for example to study the mechanisms behind drug resistance. In another application, NGS is being used to investigate genomic sequence diversity in bacterial cell populations. NGS is transforming these metagenomic studies by generating DNA sequence data from the bacterial community as a whole, which can then be used to identify the individual bacteria types present, for example within the human gut. A recent study investigated the microbiota community of the human gut and uncovered a gene set some 150 times larger than that in humans14. Metagenomic studies of case and control subjects are also allowing insight into the role of bacterial diversity in disease; for example aiding drug discovery by attempting to understand bacterial cell population diversity and its role in inflammatory bowel disease and Crohn’s disease phenotypes15.
NGS has also been successfully used in target identification for anitibacterials. In one study of Mycobacterium tuberculosis, a com – pound was identified which was effective in a whole cell assay but the mechanism of action was unknown. Following generation of resistant mutants, NGS was used to sequence both sensitive and resistant strains which led to the identification of mutations in a gene present in the resistant but not the sensitive strains. Subsequent complementation assays identified this gene as the target of the compound16.
RNA studies using RNA-Seq
Insights into disease processes and compound mechanism of action can be gained by the study of RNA; both mRNA and potentially small RNAs such as microRNA (miRNA).
Transcriptomics is the qualitative and quantitative study of RNA expression, which focuses on the differential regulation of genes. Gene expression measurements can potentially be used in disease staging, target validation, or as disease or pharmacodynamic biomarkers. NGS RNA sequencing (RNA-Seq) has enriched transcriptome studies due to its ability to generate large and detailed datasets17. In addition to accurate measurement of gene expression levels, the technology also provides high resolution information about transcript splice variation.
RNA-Seq is also capable of detecting transcripts created from gene fusion events, which have been observed in tumour cells. For example, a study by Levin et al. used RNA-Seq to sequence a tumour transcriptome. The sensitivity of sequencing with RNA-Seq enabled the detection of low abundance transcripts. As a result, novel splicing and gene fusions were reliably identified in addition to the quantification of gene expression18. This application of NGS could be used to identify such fusion transcripts with a view to developing compounds which selectively target the fusion protein product; a pharmacological approach which was taken in the development of Gleevec to target the Bcr-Abl fusion protein19.
A further application of RNA-Seq is small RNA profiling, in which known non-coding RNAs and novel RNA sequences can be detected20. Small RNA profiling encompasses a number of different classes of RNAs, which include microRNAs (miRNAs) and small interfering RNAs (siRNAs)21. Each of these non-coding RNAs is involved in the regulation of gene expression. The average length of a mature miRNA is 22 nucleotides, which makes the NGS an ideal platform to profile miRNAs due to the short read nature of the sequencing process22. Currently, 1424 human miRNA species have been discovered (miRBase release 17.0, www.mirbase.org) in a variety of tissues and bodily fluids23, including carcinoma24, embryonic stem cells25, plasma/serum, saliva, urine and blood. The numerous sources of miRNAs and their stability make them potential candidates for biomarkers. For example, Liu et al. created a miRNA profile using NGS to discover a group of five serum miRNAs that are differentially expressed in gastric cancer patients26. The discovery of such biomarkers could find application in the early diagnosis of disease26 and perhaps also in sub-grouping subjects in clinical trials based upon their miRNA profiles. Accurate miRNA profiling using NGS still has some challenges, including for example discerning closely related miRNA sequence variants called isomiRs22,25.
Applications in protein science
Although the majority of NGS applications described to date have centred on DNA and RNA, an emerging area of application is in the study of proteins. One such area is epigenetics, which is defined as the study of heritable changes in phenotype and effects on gene expression by mechanisms other than alterations in the DNA sequence. Examples include post translational modifications of histone proteins such as methylation and acetylation, or methylation of DNA bases, many of which are implicated in the control of gene expression13.
One novel application of NGS called CHIP-seq aims to monitor such changes. In the CHIP-seq process NGS chromatin immunoprecipitation (CHIP) together with large scale NGS sequencing is used to produce whole genome maps of the location of transcription factors or modified histones of interest27. ChIP-seq studies of histone modifications have also increased knowledge around gene regulation in cancer research, especially when comparing normal to tumour cells. NGS is providing a substantial amount of new data in the field of epigenetics, which may lead to the identification of new drug targets in oncology and other therapy areas28.
An intriguing new application of NGS in protein analysis is ribosome profiling. This method identifies the positions of active ribosomes that are bound to a target mRNA, providing a whole cell view of protein translation29. The NGS process captures ribosome protected mRNA, sequences the ribosome binding sites and maps the sequence reads back to the human genome, identifying the proteins which are being translated. Ingolia et al. showed how ribosome profiling in yeast can be used to determine the translational regulation of gene expression29. This method could potentially be applied to the study of tumours, as protein translation is often deregulated in these cells.
Conclusions and discussions
NGS is already finding application in many stages of the drug discovery process, from target identification through to personalised medicine and is also improving our knowledge of complex biological systems and diseases. The technology offers significant benefits when compared to conventional Sanger sequencing, due to its much lower cost-per-base and sub – stantial data sets generating billions of bases of data in a single sequencing run. The technology can carry out a range of applications all on a single platform that extend across DNA, RNA and protein studies. Finally, the emerging third generation sequencing technologies promise even larger datasets at reduced cost and the potential to sequence from small amounts of starting material which could benefit clinical studies. It is likely that DNA sequencing technologies will continue to develop at a rapid pace in the coming years, yielding novel approaches and applications in the drug discovery process.
References
1. Maxam, A.M. and Gilbert, W. (1977) A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74, 560-564
2. Sanger, F. et al. (1977) DNA sequencing with chainterminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463-5467
3. Bhullar, B. (2010) The Sequencing Revolution: enabling personal genomics and personalised medicine. European Pharmaceutical Review 5, 49-52
4. Lander, E.S. et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860-921
5. Ronaghi, M. et al. (1998) A sequencing method based on real-time pyrophosphate. Science 281, 363-365
6. Ansorge, W.J. (2009) Next-generation DNA sequencing techniques. N. Biotechnol. 25, 195-203 7. Margulies, M. et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376-380
8. Su, Z. et al. (2011) Next-generation sequencing and its applications in molecular diagnostics. Expert Rev. Mol. Diagn. 11, 333-343
9. Rothberg, J.M. and Leamon, J.H. (2008) The development and impact of 454 sequencing. Nat. Biotechnol. 26, 1117-1124
10. Metzker, M.L. (2010) Sequencing technologies – the next generation. Nat. Rev. Genet. 11, 31-46
11. Bentley, D.R. et al. (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53-59
12. Schadt, E.E. et al. (2010) A window into third-generation sequencing. Hum. Mol. Genet. 19, R227-R240
13. Mardis, E.R. (2008) The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133-141
14. Qin, J. et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59-67
15. Willing, B.P. et al. (2010) A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844-1854
16. Andries, K. et al. (2005) A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223-227
17. Wang, Z. et al. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57-63
18. Levin, J.Z. et al. (2009) Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 10, R115
19. Deininger, M.W. and Druker, B.J. (2003) Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol. Rev. 55, 401-423
20. Ozsolak, F. and Milos, P.M. (2011) RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87-98
21. Ghildiyal, M. and Zamore, P.D. (2009) Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94-108
22. Lee, L.W. et al. (2010) Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 16, 2170-2180
23. Weber, J.A. et al. (2010) The microRNA spectrum in 12 bodily fluids. Clin. Chem. 56, 1733-1741
24. Mizuguchi, Y. et al. (2011) Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One 6, e15304
25. Morin, R.D. et al. (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610-621
26. Liu, R. et al. (2011) A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur. J. Cancer 47, 784-791
27. Park, P.J. (2009) ChIP–seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669-680
28. Neff, T. and Armstrong, S.A. (2009) Chromatin maps, histone modifications and leukemia. Leukemia 23, 1243-1251
29. Ingolia, N.T. et al. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218-223
Issue
Related topics
DNA, Drug Discovery, Next Generation Sequencing, Proteins, RNA