Segregation of molecular mechanisms of genotoxicity and carcinogenicity across human, yeast and Salmonella species
Posted: 22 February 2010 | Willem Schoonen, Department of Toxicology and Drug Disposition, MSD, The Netherlands, Walter Westerink, Department of Toxicology and Drug Disposition, MSD, The Netherlands and Jean Horbach, Department of Toxicology and Drug Disposition, MSD, The Netherlands
Screening assays for in vitro toxicity are the way to reduce the attrition rates in the preclinical development of new drugs. Here a test battery is presented for screening of genotoxic and carcinogenic compounds by means of VitotoxTM, RadarScreen, and four human liver HepG2 cell lines with two different promoters as well as responsive element (RE) settings in combination with a luminescent read-out. The VitotoxTM assay in Salmonella is a substitute for the Ames test and the RadarScreen assay for in vitro clastogenicity. Moreover, HepG2 assays with RAD51C and Cystatin A promoters, and p53-RE are more predictive for in vivo clastogenicity. The Nrf2-RE can be used for analysis of reactive oxygen species production. The validity of this battery is checked for 62 compounds of an ECVAM list for genotoxicity and for 190 other references or in house drugs.

Figure 1
Screening assays for in vitro toxicity are the way to reduce the attrition rates in the preclinical development of new drugs. Here a test battery is presented for screening of genotoxic and carcinogenic compounds by means of VitotoxTM, RadarScreen, and four human liver HepG2 cell lines with two different promoters as well as responsive element (RE) settings in combination with a luminescent read-out. The VitotoxTM assay in Salmonella is a substitute for the Ames test and the RadarScreen assay for in vitro clastogenicity. Moreover, HepG2 assays with RAD51C and Cystatin A promoters, and p53-RE are more predictive for in vivo clastogenicity. The Nrf2-RE can be used for analysis of reactive oxygen species production. The validity of this battery is checked for 62 compounds of an ECVAM list for genotoxicity and for 190 other references or in house drugs.
Within the pharmaceutical industry in vitro high throughput screening (HTS) with nuclear and membrane receptors, as well as ion channel and enzyme screens has been well accepted in the early discovery phase. However, in vitro screening within toxicology remains restricted towards Ames, chromosomal aberration, micronuclei, phototoxicity and eye irritation tests. Cumbersome is that these tests are time consuming, cost expensive, require relatively large amounts of compound and are performed as low throughput screens. Moreover, the predictivity of the in vitro chromosomal aberration and micronuclei tests is far from optimal for in vivo genotoxicity and carcinogenicity. As a consequence, these assays are under heavy and ongoing debate within the ICH guidelines (ICH guideline S2B). This is especially based on the high amount of false positives of these classical in vitro tests. Due to new knowledge and insights in molecular mechanisms, novel luciferase-based in vitro assays for the assessment of genotoxicity and carcinogenicity are proposed. These differ from the current assays in that they do not measure end points of DNA damage but rather the induction of molecular mechanisms or pathways. The very high sensitivity (75 to 90%), medium to high throughput (16 to 80 compounds), low amount of compound required (3 to 10 mg), short assay time (3 to 48 h) and easy luminescence read-out of these assays make it feasible to identify genotoxic, i.e. mutagenic and clastogenic compounds, in a concentration range of 10-8 up to 10-3 M in the ‘early’ discovery phase, long before the final approval for preclinical toxicity testing of new drugs. This pro-reactive attitude will give the opportunity to modify potential drugs in their chemical constitution by elimination of the unwanted side effects and maintenance of their pharmaceutical potency. Such an approach can reduce the attrition rates tremendously. Within the former Organon, currently Schering-Plough, with a portfolio focused on male and female contraception, fertility and hormone replacement therapy, a high frequency of 20% positive scores on drug candidates for genotoxicity and/or carcinogenicity has historically been observed. The estrogenic, androgenic and progestagenic potential as well as the steroidal backbone of these compounds caused these positive outcomes. Therefore early identification of such unwanted side effects will largely improve the quality of the compounds to be developed, and reduce the number of animal studies with compounds with a low success rate.
The first set of luminescence assays was developed to replace the Ames mutagenicity test, in which different Salmonella typhimurium strains are used with either base-pair or frame shift mutations in genes involved in histidine synthesis, making them histidine-dependent for growth. The newly developed tests were no longer measuring these end points of gene mutation, but were focused on the activation of the SOS response system. This SOS reponse system was activated upon DNA damage and lead to higher expression levels of DNA repair genes to restore this damage. Activation of the response genes is thus a more indirect and earlier indicator of increased DNA damage. By replacing these response genes by luciferase (luc), β-galactosidase (LacZ) or alkaline phosphatase, an easy read-out for DNA damage can be obtained.
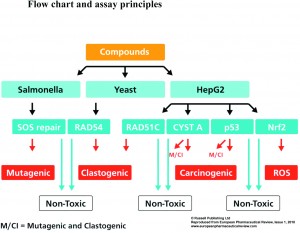

Figure 1
The SOS chromotest is one of the best known and validated assays1-3. In this test 330 chemicals were analysed, leading to predictivity, sensitivity, and selectivity scores of 86%, 79%, 100%, respectively. Later the VitotoxTM assay with S. typhimurium TA104 cells became commercially available (Thermo scientific, Finland)4,5. This SOS-response system is built with a luciferase gene, which is under transcriptional control of the recN promotor. Normally, this recN promoter is strongly repressed, but in the presence of a DNA damaging compound, the RecA regulator protein recognises the resultant free ends or mismatches in the DNA. This results in a cascade of reactions leading to derepression of the strong recN promoter. A second S. typhimurium TA104 strain constitutively expressing luciferase is used for measuring cytotoxicity and correction for false positive results. This VitotoxTM assay was evaluated with an ECVAM compound list for 20 genotoxic and 42 non-genotoxic compounds6, and 190 other reference, steroidal and in-house compounds7.
The second set of assays was developed to replace chromosomal aberration, micronuclei and COMET assays. These assays recognise double strand breaks and failures in mitotic and meiotic segregation, which is very common in eukaryotes. In yeast, the mRNA and protein expression of the RAD54 gene is clearly up-regulated, if chromosomal double strand breaks are induced by a genotoxic compound8,9. In case of GreenScreen (Gentronix, UK) and RadarScreen (reMYND, Belgium) assays, this RAD54 gene was replaced by either Green Fluorescence Protein (GFP) or LacZ. A major drawback of GFP is the autofluorescence of approximately 25% of the tested compounds at this wavelength. These effects are absent with LacZ- or luciferase-based assays. Therefore also the RadarScreen assay was evaluated with the above mentioned compound sets7.
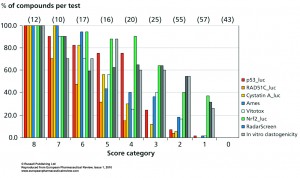

Figure 2
With these mutagenic and clastogenic cellular assays the involvement of metabolism is evaluated by the addition of an S9-fraction. This S9-fraction is a 9000 g supernatant of the liver homogenate of rats which were pre-treated for 24 hours with Aroclor 1254 or a compound cocktail that activates the Aryl hydrocarbon Receptor (AhR), Pregnane X Receptor (PXR) and the Constitutive Androstane Receptor (CAR) and thus induces the cytochrome P450 enzymes, i.e. CYP 1A1, 1A2, 3A1/2, 2B1 and 2B2. For these phase I metabolic enzymes the appropriate co-factors are added to the S9-fraction, but the influence of phase II metabolic enzymes is neglected, as co-factors for these enzymes are normally not supplied with the S9-fraction. Moreover, this metabolism is rat and not human specific, while quickly degrading or very reactive intermediates may not be able to penetrate the cellular and nuclear membrane and, therefore, will not come in close contact with the DNA. In literature, examples with a better predictivity are described, e.g. when human liver S9-fractions, primary hepatocytes or permanent cell lines are used10.
To circumvent these problems, a third set of luciferase-based reporter assays was developed with human permanent HepG2 cells. These hepatocyte-derived cells have been well characterised with respect to their capability of phase I and II metabolism11,12. Expression of mRNA and/or enzyme levels of CYP 1A1, 1A2, 3A4, UDP-glucuronosyl transferase (UGT 1A1, 1A6), sulfotransferase (SULT 1A1, 1A2, 1E1, 2A1), N-acetyltransferase (NAT1), epoxide hydrolase (EPHX1) and glutathione-S-transferase (GST 1, m1) could be identified. Therefore, these HepG2 cells were used to prepare simple read-out assays with the RAD51C and Cystatin A promoter. Activation of the RAD51C gene is also linked to chromosomal double-strand breakage, while activation of the cystatin A gene is linked to genotoxic and apoptotic processes. In line, a human lymphoblastoid cell line TK6 has been developed with a GADD45a-GFP read-out (GreenScreen HC, Gentronix), which, however, still lacks endogenous metabolism and requires rat S9 treatment13.
For the prediction of carcinogenesis an additional assay was developed, since phosphorylation of the tumour suppressor protein p53 may protect against uncontrolled cell growth, as was shown with p53 knock-out mice14. Besides that, the cleavage of the antioxidant transcription factor, the Nrf2 protein, from the Keap-1 protein under influence of reactive oxygen species (ROS) may lead to the up-regulation of genes and enzymes, like catalases, superoxide dismutases, glutathione synthetases and reductases, all involved in diminishing the amounts of ROS. This ROS activation is only indirectly linked towards genotoxicity due to the harmfull effects of ROS. The activation of p53 and Nrf2 proteins is measured by using specific responsive elements for either p53 or Nrf2 in a promoter luciferase-based HepG2 assay. Also these assays were validated with the ECVAM compound list and the 190 additional compounds. Reporter assays for p53 have also been developed by Ohno et al.15 and Sohn et al.16, while reporter assays for Nrf2 were also described by Boerboom et al.17 and Wang et al.18. However, the number of genotoxic compounds tested in these studies was rather small.
All 252 compounds were tested in the VitotoxTM and RadarScreen assays in either the presence or absence of a rat liver S9-fraction, while no S9-fraction was used in the four HepG2 reporter assays. This results either in positive or negative scores for genotoxicity or carcinogenicity (see Figure 1). The predictive values for the 20 genotoxic and 42 non-genotoxic ECVAM compounds with these tests are given in Table 1. These data show a high sensitivity for the p53_luc (85%), VitotoxTM + RadarScreen (85%), Nrf2-Luc (75%), VitotoxTM, (70%), RadarScreen (70%), and Cystatin A_luc (70%) assays, while a slightly lower sensitivity of 60% was found for the RAD51C_luc. The sensitivity of the individual scores for VitotoxTM and RadarScreen were lower than combined as the ECVAM compound list contains mutagenic and/or clastogenic compounds. The specificity for these assays varied between 81 and 95%, with an exception for Nrf2_luc (69%). Also the predictivity for these assays varied between 79 and 90%, except again for Nrf2-luc (71%). Thus with respect to the ECVAM compound list a good sensitivity, specificity and predictivity was found for these assays with exception of the lower values found with Nrf2_luc assay.
The predictive values for the other 190 compounds with these assays are given in Table 2. Here a comparison was made between VitotoxTM, RadarScreen, and the four human HepG2 assays with respectively Ames, in vitro or in vivo clastogenicity data from literature and in house analysis. The HepG2 assays were compared individually and as a combination of the three HepG2 genotox assays p53_luc, RAD51C_luc and Cystatin A_luc. The sensitivity for the VitotoxTM, RadarScreen, Human Genotox and Nrf2_luc was in the range of 74 up to 86%. Only the individual score of the RAD51C_luc assay was with 29% quite low, while the others p53_luc (71%) and Cystatin A_luc (61%) were clearly much better. The specificity varied between 81 and 94% for the VitotoxTM, p53_luc, Rad51C_luc, Cystatin A_luc, and the combined human genotox assays, while the specificity for the RadarScreen (74%) and Nrf2_luc (66%) assays was slightly lower. The predictivity for these assays varied between 76 and 91% with exception of RAD51C_luc and Nrf2_luc being respectively 59 and 71%. Overall comparison of the used human, yeast and Salmonella assays shows, that the in vitro HepG2 reporter genotox assays are a better predictor of in vivo clastogenicity, RadarScreen of in vitro clastogenicity and VitotoxTM of Ames scores. In this analysis with the human reporter assays, compounds like dimethylnitrosamine and cyclophosphamide were not identified as genotoxic most likely due to the lack of sufficient CYP2E1 or CYP2B6 activity in HepG2 cells. On the other hand, compounds like aflatoxin B1, benzo[a]pyrene, 7,12-dimethylbenzanthracene, and 2-acetylaminofluorene were clearly identified in the HepG2 cell lines, as being genotoxic. For these compounds, it is well-known, that liver phase I metabolism is essential and in favour of HepG2 cells.
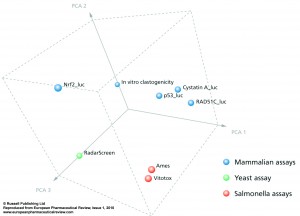

Figure 3
The advantage of these new luciferase-based assays for genotoxic and carcinogenic testing is its HTS potential. This will identify whether the effect is common to all cell lines or whether it is human, yeast or Salmonella specific. For instance, overall analysis of steroids learns that only estrogens with an aromatic A-ring in combination with a 3-hydroxy-group may be clastogenic, while many androgens and progestagens, as well as estrogens with a non-steroidal backbone or a Δ5-10 configuration, although identified as positive with the in vitro clastogenicity and RadarScreen assays, were identified as negative in these HepG2 assays. Therefore, by compound screening in all these assays, only those with positive scores in every assay will be general genotoxins, while others may appear to be human specific (curcumin and propyl gallate), or may depend on specific enzyme mechanisms, as azo- or nitro- reduction, which are only active in Salmonella. Prioritisation of compounds can be done by simple summation of all positive results on all assays (see Figure 2), by principal component analysis (see Figure 3), or by hierarchical clustering (See Figure 4) and these results can be compared with available Ames and in vitro clastogenicity data. Comparison with in vivo clastogenicity data is hampered due to the relatively low amount of in vivo clastogens in the compound set (only 70 out of 252).
In Figure 2 the percentage of positive scores per test are shown per score category for the p53_luc, Rad51C-Luc, Cystatin A-luc, Ames, VitotoxTM, Nrf2_luc, RadarScreen and in vitro clastogenicity. At score 8 all compounds are positive in all tests, while at score 7 one and at score 6 two of these tests failed for one of these compounds and so on. Thus with a score of 8, 7, 6 and 5 real genotoxic and/or carcinogenic compounds are identified. A clear reduction for the identification of positives is already seen with RAD51C_luc at score 7, 6 and 5, whereas at score 5 also the predictivity with Ames and VitotoxTM dropped sharply. At score 4, a more pronounced segregation is observed between higher scores for p53_luc, Nrf2_luc, RadarScreen and in vitro clastogenicity versus lower scores for the RAD51C-luc, Cystatin_luc, Ames and VitotoxTM assays. This becomes even more pronounced at score 3 and 2 where also the p53-luc values are reduced. At score 1, individual positive compounds were only identified with Nrf2_luc, RadarScreen and in vitro clastogenicity assays. The compounds with score 1 and 2 should be analysed with care, since they may be species specific carcinogens and/or genotoxins, they may miss the proper metabolic activation in the HepG2 assays or they are only dose dependent ROS producers, which can be prevented with radical scavengers or lower dosing. At score 0, very clean compounds are identified.
| VitotoxTM | RadarScreen | RadarScreen | HepG2 genotox reporters | HepG2 | ||||
| + Vitotox TM | p53_luc | RAD51C_luc | Cystatin A_luc | Total | Nrf2_luc | |||
| Sensitivity | 70 (14/20) | 70 (14/20) | 85 (17/20) | 85 (17/20) | 60 (12/20) | 70 (14/20) | 85 (17/20) | 75 (15/20) |
| Specificity | 93 (39/42) | 83 (35/42) | 81 (34/42) | 93 (39/42) | 95 (39/42) | 90 (38/42) | 81 (34/42) | 69 (29/42) |
| Predictivity | 85 (53/62) | 79 (49/62) | 82 (51/62) | 90 (56/62) | 82 (51/62) | 84 (52/62) | 82 (51/62) | 71 (44/62) |
Table 1 Predictive values (%) of the VitotoxTM, RadarScreen, VitotoxTM + RadarScreen, RAD51C-luc, Cystatin A-luc, p53_luc and Nrf2-luc assays with respect to the ECVAM compound list, including 20 genotoxic and 42 non-genotoxic compounds (Westerink et al., 2009a). Between brackets are the numbers of true positive or/and true negative scores followed by the total numbers of compounds tested.
In Figure 3 a principle component analysis is performed, showing that there is a very large similarity between the Ames and VitotoxTM data, which segregate largely from those of the human HepG2 genotoxicity assays with p53_luc, Cystatin A_luc, and RAD51C luc. Moreover, the in vitro clastogenicity and RadarScreen assays show a similar direction of change from the Ames/Vitotox as well as from the HepG2 genotoxicity assays. Again a clear segregation of human versus yeast is visualised. The data with the Nrf2-luc assays are completely different from these genotoxic and clastogenic mechanisms. If the available in vivo clastogenicity data of the 70 compounds are transposed into this figure, they are seen in close proximity with the human genotoxic assays.
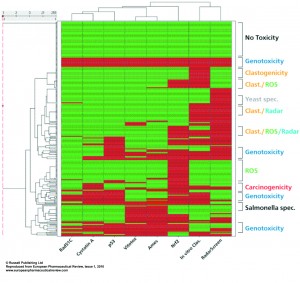

Figure 4
In Figure 4 the hierarchical clustering is shown. Also in this combination the segregation between the human genotoxic assays and VitotoxTM/Ames as well as with the Nrf2-luc, RadarScreen and in vitro clastogenicity scores becomes visible. As in Figure 2 a segregation of compounds is seen across these assays, there are categories of compounds with only positive or negative scores in all the assays. There are compounds that only show positive scores with the RadarScreen, Nrf2-luc or in vitro clastogenicity assays, while other positive scores are found in combination of the RadarScreen with either the Nrf2_luc and/or in vitro clastogenicity assays. Moreover there are compounds that are only positive in the Ames and VitotoxTM assays, being more species specific. Also from this hierarchical clustering a score category can be obtained. Although for in vivo clastogenicity comparison only a relatively low number of 70 compounds is available, these assay results correlate much better with the human genotoxic assays, than with the Salmonella or yeast assays19.
A positive genotoxic score is seen as a strong negative factor for a compound, in many cases leading to deselection of that compound. At present the VitotoxTM and (mini-) Ames tests are used in the pre-selection phase within our company. Expanding this battery with the Radarscreen and HepG2 genotoxicity assays in the early phase of LO has an important advantage in early deselection and/or ranking of the lead or in the synthesis of a chemically modified lead. It also appears that the obtained data do not always show a black or white presentation, as variation due to species specificity does occur.
| VitotoxTM | RadarScreen | HepG2 genotox reporters | HepG2 | ||||
| p53_luc | RAD51C_luc | Cystatin A_luc | total | Nrf2_luc | |||
| Sensitivity | 86 (42/49) | 77 (59/77) | 71 (27/38) | 29 (11/38) | 61 (23/38) | 74 (28/38) | 76 (29/38) |
| Specificity | 94 (90/96) | 74 (34/46) | 81 (26/32) | 94 (30/32) | 94 (30/32) | 81 (26/32) | 66 (21/32) |
| Predictivity | 91 (132/145) | 76 (93/123) | 76 (53/70) | 59 (41/70) | 76 (53/70) | 77 (54/70) | 71 (50/70) |
Table 2 Predictive values (%) of the VitotoxTM, RadarScreen, RAD51C_luc, Cystatin A_luc, p53_luc and Nrf2_luc assays with respect to the 190 additional compounds in comparison with Ames, in vitro (bold) and in vivo (underlined) clastogenicity data (Westerink et al., 2009b). Between brackets are the numbers of true positive or/and true negative scores followed by the total numbers of compounds tested.
Conclusion
A battery of novel in vitro assays for genotoxicity and carcinogenesis is available and is ready for implementation. These novel methods will be a valuable addition to the current regulatory required studies, because they mimic the in vivo biological complexity based on the segregation of molecular mechanisms. So, when specific mechanistic issues are included in in vitro methods, this will play an important role in preclinical development and in the interpretation of the human relevance. Thus in vitro toxicology in drug development can definitely lead to a lower attrition rate and reduction in animal use.
References
- V. Mersch-Sundermann, U. Schneider, G. Klopman, H.S. Rosenkranz. SOS induction in Escherichia coli and Salmonella mutagenicity: a comparison using 330 compounds, Mutagenesis Issue 9, pp. 205-224, (1994).
- P. Quillardet, C. de Bellecombe, M. Hofnung. The SOS Chromotest, a colorimetric bacterial assay for genotoxins: validation study with 83 compounds, Mutat. Res. Issue 147, pp. 79-95, (1985).
- P. Quillardet, M. Hofnung. The SOS chromotest: a review, Mutat. Res. Issue 297, pp. 235-279, (1993).
- D. Lelie van der, L. Regniers, B. Borremans, A. Provoost, L. Verschaeve. The VITOTOX test, an SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics, Mutat. Res. Issue 389, pp. 279-290, (1997).
- L. Verschaeve, J. Van Gompel, L. Thilemans, L. Regniers, P. Vanparys, D. van der Lelie. VITOTOX bacterial genotoxicity and toxicity test for the rapid screening of chemicals, Environ. Mol. Mutagen. Issue 33, pp. 240-248, (1999).
- D. Kirkland, P. Kasper, L. Muller, R. Corvi, G. Speit. Recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests: a follow-up to an ECVAM workshop, Mutat. Res. Issue 653, pp. 99-108, (2008).
- W.M.A. Westerink, J.C.R. Stevenson, A. Lauwers, G. Griffioen, G.J. Horbach and W.G.E.J. Schoonen. Evaluation of the VitotoxTM and RadarScreen assays for the rapid assessment of genotoxicity in the early research phase of drug development. Mutat. Res. Issue Apr 22. [Epub ahead of print], pp., (2009a).
- O.M. Mazina, M.J. Rossi, N.H. Thomaa, A.V. Mazin. Interactions of human rad54 protein with branched DNA molecules, J. Biol. Chem. Issue 282, pp. 21068-21080, (2007).
- G.M. Cole, D. Schild, S.T. Lovett, R.K. Mortimer. Regulation of Rad54 and RAD52-LacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage, Mol. Cell Biol. Issue 7, pp. 1078-1084, (1987).
- D.Kirkland, S.Pfuhler, D.Tweats, M.Aardema, R.Corvi, F.Darroudi, A.Elhajouji, H.Glatt, P.Hastwell, M.Hayashi, P.Kasper, S.Kirchner, A.Lynch, D.Marzin, D.Maurici, J.R.Meunier, L.Muller, G.Nohynek, J.Parry, E.Parry, V.Thybaud, R.Tice, J.van Benthem, P.Vanparys, P.White. How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: Report of an ECVAM Workshop, Mutat. Res. Issue, 628, pp. 31-55, (2007).
- W.M. Westerink, and W.G. Schoonen. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells, Toxicol. in vitro Issue 21, pp. 1581-1591, (2007a).
- W.M. Westerink, and W.G. Schoonen. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells, Toxicol. in vitro Issue 21, pp. 1592-1602, (2007b).
- P.W. Hastwell, L.L. Chai, K.J. Roberts, T.W. Webster, J.S. Harvey, R.W. Rees, R.M. Walmsley. High-specificity and high-sensitivity genotoxicity assessment in a human cell line: validation of the GreenScreen HC GADD45a-GFP genotoxicity assay, Mutat. Res. Issue 607, pp. 160-175, (2006).
- L.A. Donehower. The p53-deficient mouse: a model for basic and applied cancer studies. Seminars in Cancer Biology, Issue 7, pp. 269-278, (1996).
- K. Ohno, K. Ishihata, Y. Tanaka-Azuma, T. Yamada. A genotoxicity test system based on p53R2 gene expression in human cells: Assessment of its reactivity to various classes of genotoxic chemicals, Mutat. Res. Issue 656, pp. 27-35, (2008).
- T.A.Sohn, R.Bansal, G.H.Su, K.M.Murphy, S.E.Kern. High-throughput measurement of the Tp53 response to anticancer drugs and random compounds using a stably integrated Tp53-responsive luciferase reporter, Carcinogenesis Issue 23, pp. 949-957, (2002).
- A.M.Boerboom, M.Vermeulen, W.H.van der, B.I.Bremer, Y.Y.Lee-Hilz, E.Kampman, P.J.van Bladeren, I.M.Rietjens, J.M.Aarts. Newly constructed stable reporter cell lines for mechanistic studies on electrophile-responsive element-mediated gene expression reveal a role for flavonoid planarity, Biochem. Pharmacol. Issue 72, pp. 217-226, (2006).
- X.J.Wang, J.D.Hayes, C.R.Wolf. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents, Cancer Res. Issue 66, pp. 10983-10994, (2006).
- W.M.A. Westerink, J.C.R. Stevenson, G.J. Horbach and W.G.E.J. Schoonen. The development of p53, RAD51C, Cystatin A, and Nrf2 luciferase reporter assays in metabolic competent HepG2 cells for the mechanistic genotoxicity and oxidative stress assessment in the early phase of the research. Mutat. Res. Submitted (2009b).
About the authors
Willem Schoonen
Willem Schoonen studied biology at the University of Utrecht, where he graduated for his PhD in 1987 on steroid pheromones in fish. From 1988-1990 he worked at the Free University of Amsterdam on hyperoxic cellular toxicity. From 1990-2007 he worked at N.V. Organon, which merged in 2008 into Schering-Plough and in 2010 into MSD. He worked in the field of nuclear receptors, endocrinology, pharmacology and in vitro toxicology and toxicogenomics.
Walter Westerink
Walter Westerink studied medical biochemistry at the Hogeschool Enschede and acquired his master degree in molecular life sciences in 2007. From 1997-2007 he was employee at N.V. Organon, which merged in 2008 into Schering-Plough and in 2010 into MSD. He worked in the field of inflammation, in vitro toxicology and toxicogenomics.
Jean Horbach
Jean Horbach studied biochemistry at the University of Leiden and did his PhD at the University of Maastricht, the Netherlands. From 1996-2007 he was employee at N.V. Organon, which merged in 2008 into Schering-Plough and in 2010 into MSD. He worked in the field of drug metabolism, genotoxicity and is head of the DMPK&Safety within the department of Toxicology and Drug Disposition.





