RNAi: an attractive choice for future therapeutics
Posted: 23 May 2007 | | No comments yet
RNA interference (RNAi) is a regulatory mechanism of most eukaryotic cells that uses small double stranded RNA (dsRNA) molecules as triggers to direct homology-dependent control of gene activity (Almeida and Allshire 2005).
RNA interference (RNAi) is a regulatory mechanism of most eukaryotic cells that uses small double stranded RNA (dsRNA) molecules as triggers to direct homology-dependent control of gene activity (Almeida and Allshire 2005).
RNA interference (RNAi) is a regulatory mechanism of most eukaryotic cells that uses small double stranded RNA (dsRNA) molecules as triggers to direct homology-dependent control of gene activity (Almeida and Allshire 2005).
Known as small interfering RNAs (siRNA) these ~21-22 bp long dsRNA molecules have characteristic 2 nucleotide 3’ overhangs that allows them to be recognised by the enzymatic machinery of RNAi that eventually leads to homology-dependent degradation of the target mRNA. In mammalian cells siRNAs are produced from cleavage of longer dsRNA precursors by the RNaseIII endonuclease Dicer (Zhang et al. 2004). Dicer is complexed with two RNA binding proteins; the TAR-RNA binding protein (TRBP) and PACT, which are involved in the hand off of siRNAs to the RNA-induced silencing complex (RISC)(Lee et al. 2006). The core components of RISC are the Argonaute (Ago) family members, In humans there are eight members of this family but only Ago-2 possesses an active catalytic domain for cleavage activity (Liu et al. 2004; Meister et al. 2004). While siRNAs loaded into RISC are double-stranded, Ago-2 cleaves and releases the “passenger” strand leading to an activated form of RISC with a single-stranded “guide” RNA molecule that directs the specificity of the target recognition by intermolecular base pairing (Tang 2005). Rules that govern selectivity of strand loading into RISC are based upon differential thermodynamic stabilities of the ends of the siRNAs (Khvorova et al. 2003; Schwarz et al. 2003). The less thermodynamically stable end is favoured for binding to the PIWI domain of Ago-2.
MicroRNAs
An important arm of RNA interference involves the micro RNAs; these are endogenous duplexes that regulate gene expression post-transcriptionally by complexing with RISC and binding to the 3’ UTRs of target sequences via short stretches of homology termed the “seed sequences” (Bartel 2004; Bartel and Chen 2004). The primary mechanism of action of miRNAs is translational repression, although this can be accompanied by message degradation (Bagga et al. 2005). The miRNA duplexes possess incomplete Watson-Crick base pairing and the antisense strand cannot be chosen by cleavage of the passenger strand as for siRNAs, therefore the antisense strand must be chosen by an alternative mechanism (Gregory et al. 2005; Matranga et al. 2005; Leuschner et al. 2006).
MicroRNAs are endogenous substrates for the RNAi machinery. They are initially expressed as long primary transcripts (pri-miRNAs), which are processed within the nucleus into 60-70 bp hairpins by the microprocessor complex which consists of Drosha-DGCR8 (Lee et al. 2003) (Han et al. 2004). The pre-miRNAs are further processed in the cytoplasm by Dicer and one of the two strands is loaded into RISC, presumably via interaction with PACT(Lee et al. 2006). Importantly, it is possible to exploit this native gene silencing pathway for regulation of gene(s) of choice. If the siRNA effector is delivered to the cell it will “activate” RISC, resulting in potent and specific silencing of the targeted mRNA. Due to the potency and selectivity of RNAi, it has become the methodology of choice for silencing specific gene expression in mammalian cells.
Control of disease-associated genes makes RNAi an attractive choice for future therapeutics. Basically every human disease caused by activity from one or a few genes should be amenable for RNAi-based intervention. This list includes cancer, autoimmune diseases, dominant genetic disorders and viral infections. RNAi can be triggered by two different pathways:
- A RNA-based approach where synthetic effector siRNAs are delivered by various carriers to target cells as preformed 21-27 base duplexes; or
- via DNA-based strategies in which the siRNA effectors are produced by intracellular processing of longer RNA hairpin transcripts (reviewed in Scherer and Rossi 2003; Hannon and Rossi 2004). The latter approach is primarily based on nuclear synthesis of short hairpin RNAs (shRNAs) that are transported to the cytoplasm via the miRNA export pathway and are processed into siRNAs by Dicer. While direct use of synthetic siRNA effectors is simple and usually results in potent gene silencing, the effect is transient. In contrast, insertion of shRNA expression cassettes into target cells, usually via viral vectors, results in long term down regulation of the target with a single administration of the vector/shRNA.
Regardless of which method for triggering RNAi is chosen, the first step in using either siRNAs or expressed hairpins involves determination of the optimal target site-siRNA combinations. There are two major components in siRNA PTGS targeting:
- Target sequence and local structure
- siRNA thermodynamic end properties and internal sequence motifs.
We and many other groups have developed algorithms to analyse both the target accessibility and the thermodynamic end stabilities of siRNAs to target sequences with favourable thermodynamic end properties. Each algorithm has utility for choosing siRNAs. Our algorithm (Heale et al. 2005) is freely available via the link http://coh-mfold.sdsc.edu/. The sequence to be examined is entered in a FASTA format and the algorithm will rank potential siRNA targets from best to worst. In addition, the program calculates the thermodynamic end properties of the potential siRNA for these targets. The differential between 5’ end stability and 3’ end stability is calculated, allowing a quick evaluation of which strand may be preferentially incorporated into RISC. In this type of analysis one needs to evaluate the siRNA/target combination by looking at both accessibility index and siRNA properties. An accessible target that has an siRNA with poor thermodynamic end properties should be avoided, whereas a lesser ranked target with a good siRNA is a better choice.
Figure 1 depicts the relative predictive values of secondary structures versus siRNA properties for knockdown of a collection of published targets. Of note is the fact that the most effective predictors of siRNA efficacy are combining the target site accessibility with the siRNA thermodynamic end properties.
It is generally useful to pick at least three potential siRNAs for further testing. For synthetic siRNA triggers we use Dicer substrates of the structure illustrated in Figure 2, where the siRNA is produced from a longer precursor (Rose et al. 2005) which can be several fold more effective than conventional 21mers because the processing is coupled with Dicer handoff to RISC. If the approach uses shRNAs, it is important to make the shRNAs long enough to be processed by the enzyme Dicer to accurately predict the siRNA that will be produced. This means using a hairpin of greater than 23 base pairs in length. The simplest design for incorporating the optimal siRNAs into our expression constructs is to embed these in hairpins, which can be expressed from Pol III and Pol II promoters. From previous studies of Dicer substrates in cells as well as using purified recombinant human Dicer (Rose et al. 2005; Siolas et al. 2005), we know that substrates which direct Dicer to cleave a single siRNA from a precursor have a two base 3’ overhang. Dicer uses its Paz domain to enter the substrate from the 3’ overhang, and cleaves the substrate 21 bases from the 5’ end of the sense (upper) and antisense (lower) strands. We have carried out ES-Mass Spec analyses on the cleavage products of the 25/27 base asymmetric substrates, and observe only a single species of siRNA which includes the first 21 bases of the bottom strand and the first 21 bases of the sense or upper strand (Rose et al. 2005). These results provide a framework for the design of shRNAs in which the hairpins are 25-to 27 bases, with the desired siRNA comprising the first 19-21 bases of the hairpin. To ensure that the desired 5’ end of the antisense strand is produced, Pol III expression of shRNAs works best since Pol III terminates in a run of U’s, leaving short U stretches at the 3’ ends of the transcripts which serve as the Dicer entry sequence. The other strategy is to incorporate the siRNA within the context of a microRNA, such as mir30 (Zeng et al. 2002). The advantage of this particular microRNA is that its processing is well studied, and it has been successfully used as a platform for incorporation of siRNAs (Zeng et al. 2005).
The next concern for effecting RNAi is delivery of the triggers. For both synthetic siRNAs and expressed shRNAs a number of effective methods have been reported in the literature (reviewed in Kim and Rossi 2007). Whether the RNAi trigger be via delivery of synthetic siRNAs or viral vector for shRNAs, it is important to determine the efficiency of transfection or transduction. For siRNAs this is best accomplished by first using fluorescently labelled siRNAs which allow either FACS or microscopic determination of transfection efficiency. For viral vectors, it is often convenient to use a reporter gene in the vector, such as EGFP to monitor transduction efficiency. Target knockdown is measured via either analyses of mRNA levels or protein levels, which can be determined by conventional RNA and protein detection methods or via reduction of gene function. In any experimental regimen it is important to use appropriate controls to validate that the target reduction is via specific versus non-specific mechanisms. It is strongly recommended that two or more siRNAs or shRNAs to the same target be used to confirm the phenotype.
RNAi mediated transcriptional gene silencing
Recent evidence from total mammalian genome analyses of RNA expression demonstrates that RNA is transcribed throughout the genome in both sense and antisense orientations, and that there are many non-coding RNAs of varying sizes in the nucleus, many of which come from un-annotated regions of the genome, suggesting that RNAs may be involved in multiple regulatory processes in gene regulation (Carninci et al. 2005; Katayama et al. 2005; Furuno et al. 2006; Mattick and Makunin 2006; Willingham and Gingeras 2006). We and others have exploited the possible role of small RNAs in regulating transcription by targeting these RNAs to promoter regions of selected genes and have succeeded in obtaining 60-80 percent reduction in expression of the CCR5, HIV LTR, EF1A or RASSF1A promoters by targeting siRNAs to these promoter regions (Castanotto et al. 2005; Kim et al. 2006; Weinberg et al. 2006). Our lab as well as the Corey lab have established a role for RNAi components in this process in human cells (Janowski et al. 2006; Kim et al. 2006; Weinberg et al. 2006). Although our understanding of how small RNAs may direct epigenetic changes in the genome, and trigger silencing of transcription is limited, there are clear indications that we may be exploiting a natural mechanism, directed by RNAs, for changing chromatin structure.
Transcriptional gene silencing for the control of HIV infection is one of the goals of our research program. To this end the chemokine receptor CCR5, which is one of the co-receptors for HIV, represents a good target for gene silencing since expression of a functional receptor appears not to be required for normal hematopoiesis and individuals homozygous for a 32 bp deletion in this gene have greater resistance to HIV infection and a longer term progression to AIDS when infected (Arenzana-Seisdedos and Parmentier 2006). We have previously demonstrated that ribozyme mediated downregulation of CCR5 can provide some measure of protection from M-tropic HIV infection (Cagnon and Rossi 2000; Li et al. 2003), and other groups have also demonstrated that anti-CCR5 siRNAs or shRNAs are highly effective at blocking HIV infection (Martinez et al. 2002; Qin et al. 2003; Song et al. 2003; An et al. 2006). A second target for gene silencing is the HIV LTR, for which we have already defined a target and siRNA combination for silencing (Weinberg et al. 2006). With the exception of our own studies of the RASSF1A promoter, all of the experiments directing gene silencing to date have been conducted with siRNAs targeting promoter regions. Our RASSF1A studies employed shRNAs targeting promoter regions, and these were effective in both substantial silencing of the locus via histone H3K9 di-methylation and in some cells, DNA methylation of the promoter region(Castanotto et al. 2005; Kim et al. 2006). Since transcriptional gene silencing appears to be another pathway for RNA interference, it is relevant to consider this approach as a means for long term inhibition of gene expression. From the recent findings in this area, it appears that synthetic siRNAs can trigger promoter silencing via heterochromatin formation, but this is transient (Kim et al. 2006). Therefore, triggering long term transcriptional silencing may best be accomplished by using shRNA or siRNA expression systems. Whether or not RNAi mediated transcriptional gene silencing will find its way into the clinic is unknown at this time, but this is clearly a mechanism worth exploring for therapeutic intervention of gene expression.
Concluding remarks
The potential for combining transcriptional and post-transcriptional gene silencing is an exciting possibility for therapeutic application of RNAi. The challenge is to ensure that the desired silencing is specific for only the target gene(s). Delivery of siRNAs and shRNAs was initially considered to be the major roadblock to successful RNAi based therapies. This is no longer the case since many different delivery approaches have been shown to be efficacious both in cell culture and in vivo. The future for RNAi based therapeutics appears to be unlimited with the caveat that we do not yet have any clinical experience with long term RNAi based therapies. Since there are now a large number of ongoing clinical trials for RNAi, the benefits and potential pitfalls of this very powerful sequence specific mechanism for regulating gene expression will soon be forthcoming.
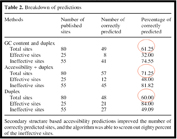

Figure 1: Parameters for choosing optimal siRNA-target combinations. The most important determinants for effective RNAi are combined target accessibility and siRNA thermodynamic end properties


Figure 2: Modeling Dicer substrates for effective generation of siRNAs from asymmetric 25/27mer Dicer substrates. The siRNA that will be generated from this Dicer substrate is indicated in bold. Dicer enters from 2 base 3’ overhang and cleaves as indicated. The blunt end of this substrate incorporates two deoxyribonucleotides at the 3’ end of the sense strand to promote Dicer entry from the two base 3’ overhang.
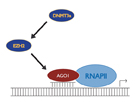

Figure 3: Model for transcriptional gene silencing by small RNAs. Small RNAs complementary to a promoter transcript are bound by an Ago family member which in turn leads to recruitment of chromatin remodeling proteins and heterochromatization.
References
Almeida, R. and Allshire, R.C. 2005. RNA silencing and genome regulation. Trends Cell Biol 15(5): 251-258.
An, D.S., Qin, F.X., Auyeung, V.C., Mao, S.H., Kung, S.K., Baltimore, D., and Chen, I.S. 2006. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther 14(4): 494-504.
Arenzana-Seisdedos, F. and Parmentier, M. 2006. Genetics of resistance to HIV infection: Role of co-receptors and co-receptor ligands. Semin Immunol 18(6): 387-403.
Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., and Pasquinelli, A.E. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122(4): 553-563.
Bai, J., Banda, N., Lee, N.S., Rossi, J., and Akkina, R. 2002. RNA-based anti-HIV-1 gene therapeutic constructs in SCID-hu mouse model. Mol Ther 6(6): 770-782.
Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2): 281-297.
Bartel, D.P. and Chen, C.Z. 2004. Micromanagers Rev of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Genet 5(5): 396-400.
Cagnon, L. and Rossi, J.J. 2000. Downregulation of the CCR5 beta-chemokine receptor and inhibition of HIV-1 infection by stable VA1-ribozyme chimeric transcripts. Antisense Nucleic Acid Drug Dev 10(4): 251-261.
Carninci, P. Kasukawa, T. Katayama, S. Gough, J. Frith, M.C. Maeda, N. Oyama, R. Ravasi, T. Lenhard, B. Wells, C. Kodzius, R. Shimokawa, K. Bajic, V.B. Brenner, S.E. Batalov, S. Forrest, A.R. Zavolan, M. Davis, M.J. Wilming, L.G. Aidinis, V. Allen, J.E. Ambesi-Impiombato, A. Apweiler, R. Aturaliya, R.N. Bailey, T.L. Bansal, M. Baxter, L. Beisel, K.W. Bersano, T. Bono, H. Chalk, A.M. Chiu, K.P. Choudhary, V. Christoffels, A. Clutterbuck, D.R. Crowe, M.L. Dalla, E. Dalrymple, B.P. de Bono, B. Della Gatta, G. di Bernardo, D. Down, T. Engstrom, P. Fagiolini, M. Faulkner, G. Fletcher, C.F. Fukushima, T. Furuno, M. Futaki, S. Gariboldi, M. Georgii-Hemming, P. Gingeras, T.R. Gojobori, T. Green, R.E. Gustincich, S. Harbers, M. Hayashi, Y. Hensch, T.K. Hirokawa, N. Hill, D. Huminiecki, L. Iacono, M. Ikeo, K. Iwama, A. Ishikawa, T. Jakt, M. Kanapin, A. Katoh, M. Kawasawa, Y. Kelso, J. Kitamura, H. Kitano, H. Kollias, G. Krishnan, S.P. Kruger, A. Kummerfeld, S.K. Kurochkin, I.V. Lareau, L.F. Lazarevic, D. Lipovich, L. Liu, J. Liuni, S. McWilliam, S. Madan Babu, M. Madera, M. Marchionni, L. Matsuda, H. Matsuzawa, S. Miki, H. Mignone, F. Miyake, S. Morris, K. Mottagui-Tabar, S. Mulder, N. Nakano, N. Nakauchi, H. Ng, P. Nilsson, R. Nishiguchi, S. Nishikawa, S. Nori, F. Ohara, O. Okazaki, Y. Orlando, V. Pang, K.C. Pavan, W.J. Pavesi, G. Pesole, G. Petrovsky, N. Piazza, S. Reed, J. Reid, J.F. Ring, B.Z. Ringwald, M. Rost, B. Ruan, Y. Salzberg, S.L. Sandelin, A. Schneider, C. Schonbach, C. Sekiguchi, K. Semple, C.A. Seno, S. Sessa, L. Sheng, Y. Shibata, Y. Shimada, H. Shimada, K. Silva, D. Sinclair, B. Sperling, S. Stupka, E. Sugiura, K. Sultana, R. Takenaka, Y. Taki, K. Tammoja, K. Tan, S.L. Tang, S. Taylor, M.S. Tegner, J. Teichmann, S.A. Ueda, H.R. van Nimwegen, E. Verardo, R. Wei, C.L. Yagi, K. Yamanishi, H. Zabarovsky, E. Zhu, S. Zimmer, A. Hide, W. Bult, C. Grimmond, S.M. Teasdale, R.D. Liu, E.T. Brusic, V. Quackenbush, J. Wahlestedt, C. Mattick, J.S. Hume, D.A. Kai, C. Sasaki, D. Tomaru, Y. Fukuda, S. Kanamori-Katayama, M. Suzuki, M. Aoki, J. Arakawa, T. Iida, J. Imamura, K. Itoh, M. Kato, T. Kawaji, H. Kawagashira, N. Kawashima, T. Kojima, M. Kondo, S. Konno, H. Nakano, K. Ninomiya, N. Nishio, T. Okada, M. Plessy, C. Shibata, K. Shiraki, T. Suzuki, S. Tagami, M. Waki, K. Watahiki, A. Okamura-Oho, Y. Suzuki, H. Kawai, J. and Hayashizaki, Y. 2005. The transcriptional landscape of the mammalian genome. Science 309(5740): 1559-1563.
Castanotto, D., Tommasi, S., Li, M., Li, H., Yanow, S., Pfeifer, G.P., and Rossi, J.J. 2005. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther 12(1): 179-183.
Dohjima, T., Lee, N.S., Li, H., Ohno, T., and Rossi, J.J. 2003. Small interfering RNAs expressed from a Pol III promoter suppress the EWS/Fli-1 transcript in an Ewing sarcoma cell line. Mol Ther 7(6): 811-816.
Furuno, M., Pang, K.C., Ninomiya, N., Fukuda, S., Frith, M.C., Bult, C., Kai, C., Kawai, J., Carninci, P., Hayashizaki, Y., Mattick, J.S., and Suzuki, H. 2006. Clusters of internally primed transcripts reveal novel long non-coding RNAs. PLoS Genet 2(4): e37.
Gregory, R.I., Chendrimada, T.P., Cooch, N., and Shiekhattar, R. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123(4): 631-640.
Gu, S., Ji, J., Kim, J.D., Yee, J.K., and Rossi, J.J. 2006. Inhibition of Infectious Human Immunodeficiency Virus Type 1 Virions via Lentiviral Vector Encoded Short Antisense RNAs. Oligonucleotides 16(4): 287-295.
Han, J., Lee, Y., Yeom, K.H., Kim, Y.K., Jin, H., and Kim, V.N. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18(24): 3016-3027.
Hannon, G.J. and Rossi, J.J. 2004. Unlocking the potential of the human genome with RNA interference. Nature 431(7006): 371-378.
Heale, B.S., Soifer, H.S., Bowers, C., and Rossi, J.J. 2005. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res 33(3): e30.
Janowski, B.A., Huffman, K.E., Schwartz, J.C., Ram, R., Nordsell, R., Shames, D.S., Minna, J.D., and Corey, D.R. 2006. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13(9): 787-792.
Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C.C., Suzuki, M., Kawai, J., Suzuki, H., Carninci, P., Hayashizaki, Y., Wells, C., Frith, M., Ravasi, T., Pang, K.C., Hallinan, J., Mattick, J., Hume, D.A., Lipovich, L., Batalov, S., Engstrom, P.G., Mizuno, Y., Faghihi, M.A., Sandelin, A., Chalk, A.M., Mottagui-Tabar, S., Liang, Z., Lenhard, B., and Wahlestedt, C. 2005. Antisense transcription in the mammalian transcriptome. Science 309(5740): 1564-1566.
Khvorova, A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs Exhibit Strand Bias. Cell 115(2): 209-216.
Kim, D.H. and Rossi, J.J. 2007. Strategies for silencing human disease using RNA interference. Nat Rev Genet 8(3): 173-184.
Kim, D.H., Villeneuve, L.M., Morris, K.V., and Rossi, J.J. 2006. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13(9): 793-797.
Lee, N.S., Dohjima, T., Bauer, G., Li, H., Li, M.J., Ehsani, A., Salvaterra, P., and Rossi, J. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol 20(5): 500-505.
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., and Kim, V.N. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956): 415-419.
Lee, Y., Hur, I., Park, S.Y., Kim, Y.K., Suh, M.R., and Kim, V.N. 2006. The role of PACT in the RNA silencing pathway. Embo J 25(3): 522-532.
Leuschner, P.J., Ameres, S.L., Kueng, S., and Martinez, J. 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7(3): 314-320.
Li, M., Li, H., and Rossi, J.J. 2006. RNAi in Combination with a Ribozyme and TAR Decoy for Treatment of HIV Infection in Hematopoietic Cell Gene Therapy. Ann N Y Acad Sci 1082: 172-179.
Li, M. and Rossi, J.J. 2005. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol Biol 309: 261-272.
Li, M.J., Bauer, G., Michienzi, A., Yee, J.K., Lee, N.S., Kim, J., Li, S., Castanotto, D., Zaia, J., and Rossi, J.J. 2003. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther 8(2): 196-206.
Li, M.J., Kim, J., Li, S., Zaia, J., Yee, J.K., Anderson, J., Akkina, R., and Rossi, J.J. 2005. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther 12(5): 900-909.
Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science.
Martinez, M.A., Gutierrez, A., Armand-Ugon, M., Blanco, J., Parera, M., Gomez, J., Clotet, B., and Este, J.A. 2002. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. Aids 16(18): 2385-2390.
Matranga, C., Tomari, Y., Shin, C., Bartel, D.P., and Zamore, P.D. 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123(4): 607-620.
Mattick, J.S. and Makunin, I.V. 2006. Non-coding RNA. Hum Mol Genet 15 Spec No 1: R17-29.
Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15(2): 185-197.
Qin, X.F., An, D.S., Chen, I.S., and Baltimore, D. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A 100(1): 183-188.
Robbins, M.A., Li, M., Leung, I., Li, H., Boyer, D.V., Song, Y., Behlke, M.A., and Rossi, J.J. 2006. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat Biotechnol 24(5): 566-571.
Robbins, M.A. and Rossi, J.J. 2005. Sensing the danger in RNA. Nat Med 11(3): 250-251.
Rose, S.D., Kim, D.H., Amarzguioui, M., Heidel, J.D., Collingwood, M.A., Davis, M.E., Rossi, J.J., and Behlke, M.A. 2005. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res 33(13): 4140-4156.
Scherer, L.J. and Rossi, J.J. 2003. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol 21(12): 1457-1465.
Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2): 199-208.
Siolas, D., Lerner, C., Burchard, J., Ge, W., Linsley, P.S., Paddison, P.J., Hannon, G.J., and Cleary, M.A. 2005. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 23(2): 227-231.
Song, E., Lee, S.K., Dykxhoorn, D.M., Novina, C., Zhang, D., Crawford, K., Cerny, J., Sharp, P.A., Lieberman, J., Manjunath, N., and Shankar, P. 2003. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol 77(13): 7174-7181.
Tang, G. 2005. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci 30(2): 106-114.
Weinberg, M.S., Villeneuve, L.M., Ehsani, A., Amarzguioui, M., Aagaard, L., Chen, Z.X., Riggs, A.D., Rossi, J.J., and Morris, K.V. 2006. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. Rna 12(2): 256-262.
Willingham, A.T. and Gingeras, T.R. 2006. TUF love for “junk” DNA. Cell 125(7): 1215-1220.
Zeng, Y., Cai, X., and Cullen, B.R. 2005. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol 392: 371-380.
Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 9(6): 1327-1333.
Zhang, H., Kolb, F.A., Jaskiewicz, L., Westhof, E., and Filipowicz, W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118(1): 57-68.
Dr. John J. Rossi
Dr. Rossi is a world-renowned expert in ribozymes (molecular scissors). Dr. Rossi’s major contributions to science have been through understanding the processing and metabolism of RNA inside the cell. One of his most notable projects is in the area of ribozyme research in AIDS. He led the research team that first suggested applying ribozymes to treat HIV. His research in molecular genetics and microbiology has earned eight patents and has served as the basis for more than 120 scientific papers. Born and raised in Washington D.C., Dr. Rossi received his bachelor’s degree from the University of New Hampshire and earned his doctorate at the University of Connecticut.
Dr. Rossi is Professor and Chair of the Division of Molecular Biology, Beckman Research Institute of the City of Hope, and Dean, Graduate School of Biological Sciences, Beckman Research Institute of the City of Hope. In the 1980s and 1990s his research focused on the mechanism of action and clinical applications of catalytic RNAs, or ribozymes. His group was the first to demonstrate that hammerhead ribozymes could be used for inhibition of HIV replication. He is the recipient of an NIH Merit award for his work on ribozymes and HIV. He has published over 200 peer reviewed articles and numerous reviews and commentaries on RNAi based therapeutics.





