Application of liquid chromatography in characterisation of lipid nanoparticle-based oligonucleotides
Posted: 28 April 2020 | Adam Socia (MSD), Andreas Abend (MSD), Yong Liu (MSD), Yuejie Zhao (MSD) | 1 comment
Duplex oligonucleotides (OGN) with lipid nanoparticles (LNPs) as delivery vehicles for different therapeutic indications are of great interest in pharma, exemplified by the recent approval of Onpattro from Alnylam Pharmaceuticals. Liquid chromatography is an essential analytical technique in the characterisation of LNPs containing OGNs. This brief review is intended to highlight the major applications of liquid chromatography in OGN purity assessment, LNP particle heterogeneity assessment and functional excipient analysis.

Potential impurities in oligonucleotides
Synthetic oligonucleotide impurities (Table 1) may arise during the manufacturing process (ie, solid phase synthesis, cleavage or deprotection steps) and can impact stability.1,2 After the annealing step of the complementary single strand OGNs to form the duplex, the residual excess of single strand is also considered an impurity.
Failure or deletion impurities are sequences that lack at least one nucleotide of single strand full length products (FLPs). Addition impurities are sequences that contain at least one additional nucleotide over FLP. If the OGN contains a triphosphate group coupled at the sequence end of the single strand, dephosphorylation impurities may arise.3-10 These hydrolytic products can form during synthesis, purification, LNP-encapsulation or storage.
Another class of impurities are FLPs with the addition of the 2´protection group tertbutyldimethylsilyl (TBDMS) as a result of incomplete deprotection.8,11 Furthermore, insufficient removal of the N-palmitoyl-4,7,10-trioxatridecane diamine, the lipid tag added to resin as part of the triphosphate chemistry, can result in tag-on impurities. Oxidation also causes impurity generation; for example, oxidation of phosphorothioate linkages. OGN purity analysis is determined mainly by denaturing anion exchange (AEX) liquid chromatography and ion‑pairing reverse-phase liquid chromatography (IP-RPLC).
Analysis of oligonucleotides by denaturing anion exchange liquid chromatography
A well-developed AEX liquid chromatography method is capable of resolving and quantitating the majority of those impurities discussed above.1,12,13 Denaturing chromatography conditions, such as elevated temperature, high pH and significant amounts of organic modifier (such as 25 percent acetonitrile, v/v) are required/ used to disrupt the duplex to form single strands.The negatively charged oligonucleotides are initially retained by the cationic stationary phase and then eluted by increasing the counter anion salt concentration in the mobile phase. This methodology is especially effective for determining the dephosphorylation products for OGNs containing triphosphate end groups and addition/deletion impurities of FLPs (ie, N-1/N+1).
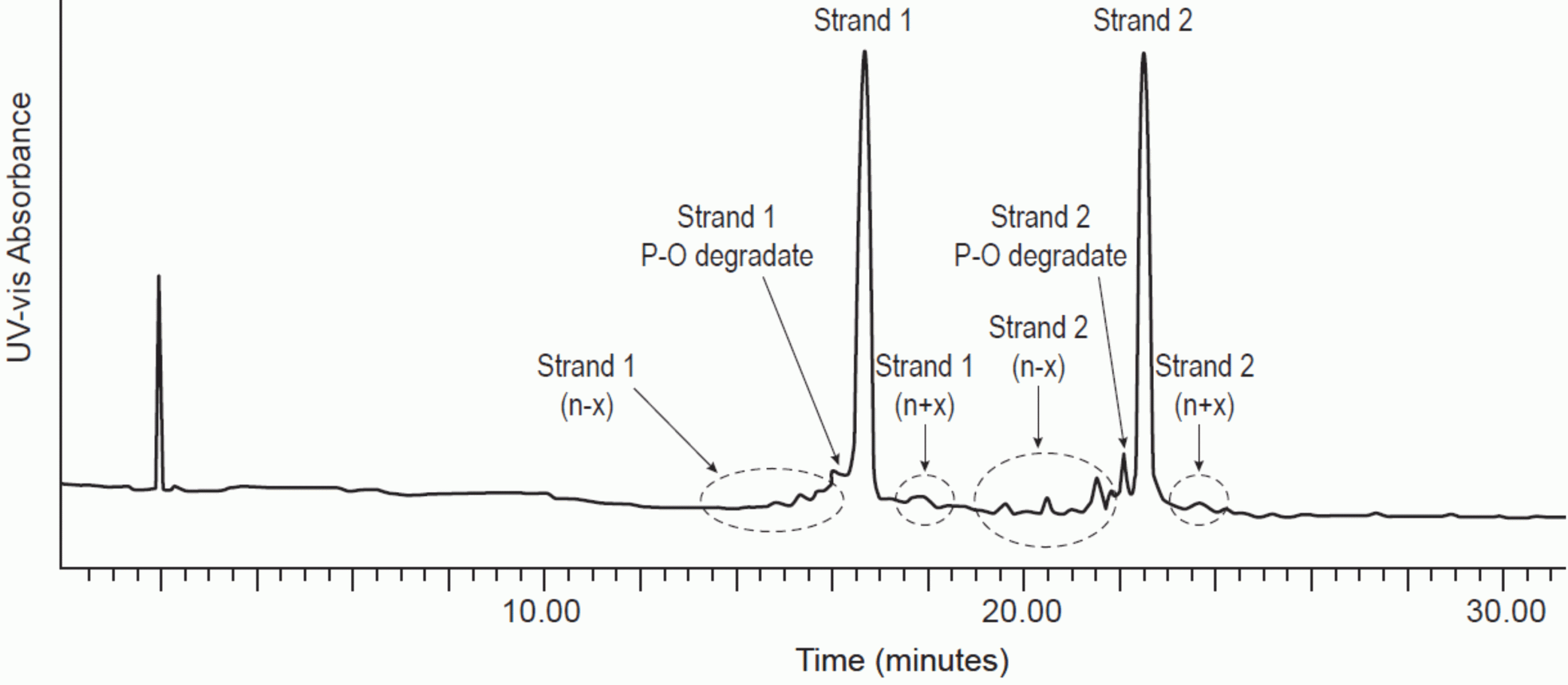

Figure 1: Denaturing IP-RPLC separation of double stranded RNA.
Mobile phase A: 16mM TEA, 400mM HFIP, pH 7.9. Mobile phase B: Methanol
Gradient: from 17 to 25 percent B over 30 minutes. C18 column, 2.1 x 300mm. 1.7μm particle size
Column temperature: 80°C. UV detection: 260nm. Flow rate: 0.6mL/min
Figures used with permission from original authors.31
The selectivity of the AEX method can be altered by adjusting the pH of the mobile phase. At a neutral pH, the retention mechanism arises from the charges on the phospho-sugar backbone and retention is relative to the length of the oligonucleotide only. However, at higher pH, the guanine, thymine and uracil-containing nucleotides can become anionic and retention will be dependent on a combination of the ionised guanine, thymine and uracil content and the oligonucleotide length, providing a way to alter selectivity.13
Due to the neutral-to-high pH mobile phases used in AEX analysis of oligonucleotides, the use of pH-stable stationary phases, such as polymeric or polymer-silica hybrid-based columns, is required.
Analysis of oligonucleotides by ion-pairing reverse-phase liquid chromatography
Table 1 Example of structures of a 5’ triphosphate synthetic single stranded oligonucleotide and its impurities.AEX, however, has two major drawbacks: limited separation power for compounds with similar affinity to ion exchangers and incompatibility with mass spectrometers (MS) due to the high salt content in mobile phase. IP-RPLC methods can be applied to address these challenges, including the analysis of single strands and duplexes, either in the denatured or native state.14-19 OGNs are highly negatively charged at neutral-to-basic pH. Therefore, cationic reagents such as triethylammonium acetate (TEAA) or triethyl amine (TEA) are generally used to ion pair with OGNs, which allows the retention of OGNs on alkyl-based “reverse-phase” stationary phases. Like traditional reverse-phase analysis, elution time is proportional to the amount of organic modifier used in the mobile phase and is related to the analyte hydrophobicity.
Component | Representative structure |
5’ Triphosphate single stranded RNA | 3’-AAAUUUGGGCCC-PPP-5’ |
“N-1” deletion (3’) | 3’-_AAUUUGGGCCC-PPP-5’ |
“N+1” addition (3’) | 3’-GAAAUUUGGGCCC-PPP-5’ |
5’ Diphosphate single stranded RNA | 3’-AAAUUUGGGCCC-PP_-5’ |
5’ Monophosphate single stranded RNA | 3’-AAAUUUGGGCCC-P__-5’ |
5’ De-phosphorolated single stranded RNA | 3’-AAAUUUGGGCCC___-5’ |
Table 1: Example of structures of a 5’ triphosphate synthetic single stranded oligonucleotide and its impurities
The addition of hexafluoroisopropanol (HFIP) to the mobile phase enables the use of a much lower amount of TEA (15mM versus 100mM) while achieving a similar ion pairing effect to TEAA. This can significantly enhance electrospray MS signal intensity, making the combination of HFIP and TEA preferred for LC-MS analysis of OGNs.20 An example of a denaturing IP-RPLC separation of OGNs is shown in Figure 1.
Denaturing, if desired, can be achieved through sample pretreatment with chemical agents or during analysis with mobile phase composition and column temperature. For instance, a higher column temperature, such as 60°C, will cause the melting of the duplex into its single strand components, therefore allowing the analysis of single strand impurities. On the other hand, analysis at ambient conditions can preserve the duplex and allow quantitation of excess single strands similar to SEC.
Analysis of oligonucleotides containing LNPs by size exclusion chromatography
The self-assembly of oligo-encapsulated LNPs based on a rapid precipitation process can result in inherent polydispersity in LNP size and composition.21 Dynamic light scattering (DLS) is routinely used to estimate the hydrodynamic size and distribution of these nanometre-sized LNPs. However, the intensity of the scattered light is highly sensitive to particle size, potentially resulting in biased (intensity) distribution results. As a bulk measurement, the size distribution result from DLS is of lower resolution compared with other methods.
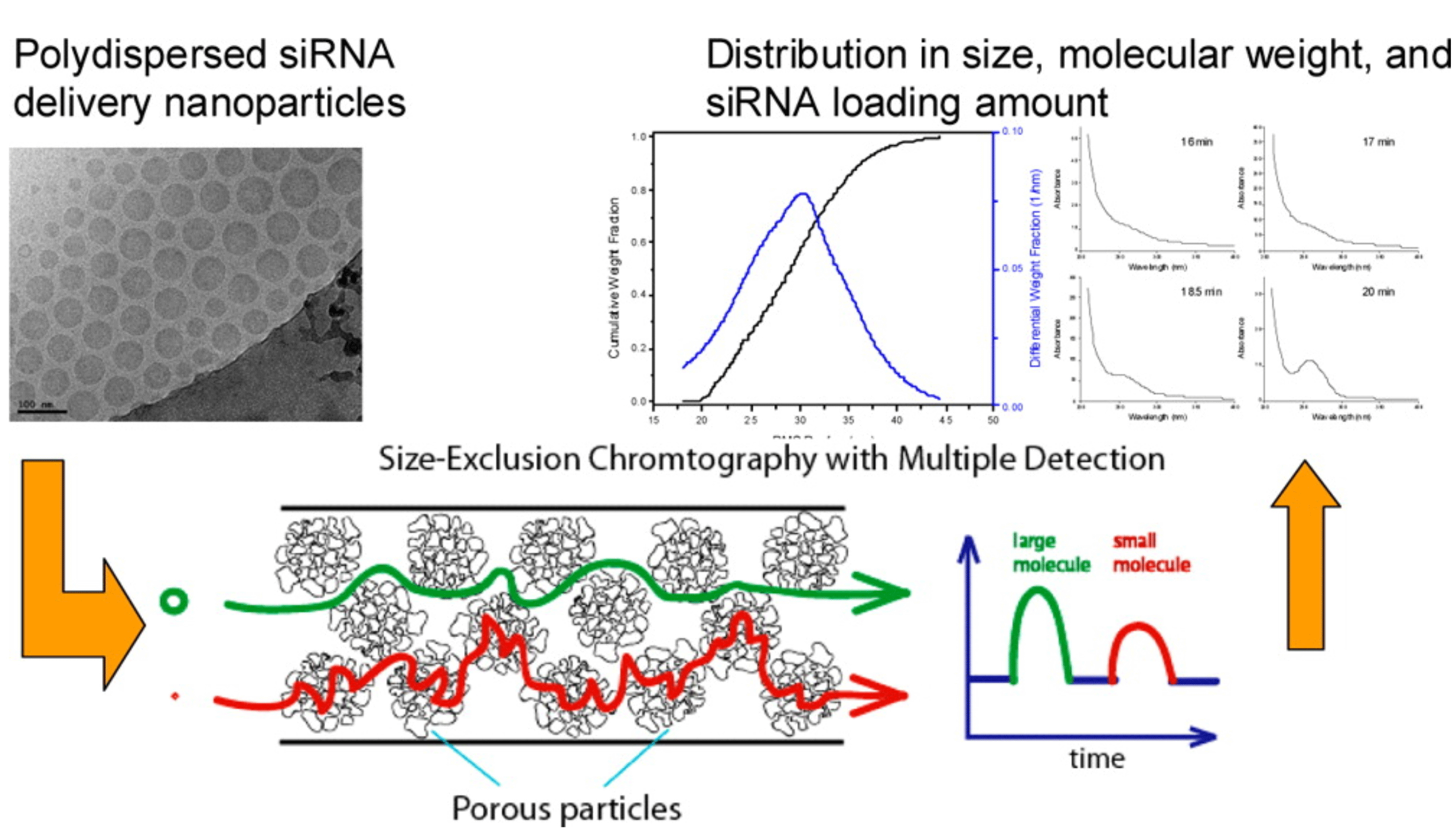

Figure 2: SEC-MALS-UV/RI characterisation of an siRNA LNP. In a single measurement, size, heterogeneity, cargo loading and MW
distribution were acquired.
Figures used with permission from original authors.24
Size exclusion chromatography (SEC) coupled with different/multiple detectors is an orthogonal tool to address the drawback of DLS. SEC columns with suitable pore sizes are selected through column screening to accommodate the size of LNPs.22 Non-denaturing conditions, which employ mild aqueous salt solutions and ambient temperatures, ensure the structure and distribution of LNPs remain unperturbed during the SEC analysis. Once a desired separation has been achieved and a non-denaturing solution condition applied, the chromatographic peak elution order from SEC can be correlated to the hydrodynamic diameters of the LNPs. The degree of polydispersity of LNPs can be roughly estimated by the peak shape. From the UV response at 260nm, one can calculate the relative loading of oligonucleotide cargo. More importantly, information regarding molecular weight distribution (MWD) and the dimensions based on their RMS radius of LNP can be obtained when a multi-angle light scattering (MALS) detector is coupled with SEC.23
Analysis of functional lipid excipients
An online SEC-MALS-UV/RI setup has been developed to provide high resolution analysis of LNPs containing siRNA. In a single measurement, size heterogeneity, cargo loading and MW distribution were acquired (Figure 2).24 Furthermore, additional understanding of the batch heterogeneity was also obtained by analysing the fractions collected from SEC separation.25 Finally, at non-denaturing conditions, SEC has also been used as an assay method for intact RNA and DNA duplexes to evaluate duplex stability during different stressed conditions.26
Another important aspect of oligo-containing LNP analysis is the characterisation of functional lipid excipients. LNPs are commonly comprised of four lipid components: a cationic lipid to encapsulate the negatively charged oligonucleotide; a zwitterionic phospholipid such as 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC); a neutral lipid such as cholesterol; and a polyethylene glycol-lipid (PEG-lipid).27,28
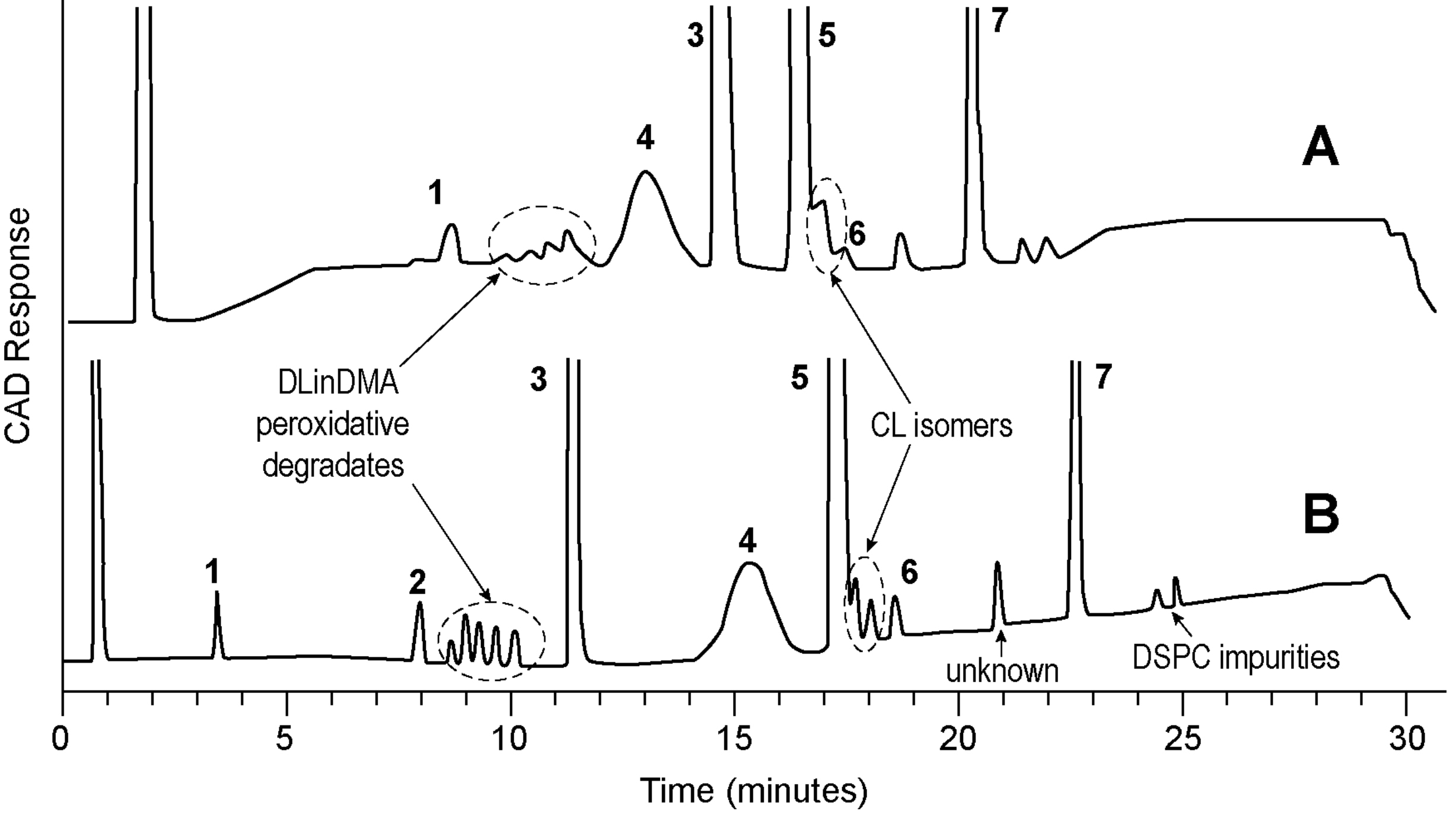

Figure 3: Comparison of HPLC-CAD (A) and UPLC-CAD (B) chromatogram of an siRNA LNP.
Mobile phase A: 0.1% TFA in water. Mobile phase B: 0.1% TFA in THF.
Gradient: from 50 to 75 percent B over 30 minutes.
C18 column, HPLC: 4.6x150mm. 3μm particle size; UPLC: 2.1x150mm. 1.7μm particle size. Column temperature: 60°C.
Peaks: 1, lyso-PC; 2, Stearic acid; 3, Cholesterol; 4, PEG-lipid; 5, cationic lipid; 6, cationic lipid N-oxide; 7, DSPC.
Figures used with permission from original authors.31
The primary degradation pathways of these lipids were reported. For instance, the amino-containing cationic lipid is susceptible to oxidation, forming an N-oxide degradant.29 The ester-containing lipids, including the phospholipid and PEG-lipids, can hydrolyse over time leading to the formation of fatty acids and lysophospholipids.30 These degradants can cause modification of LNP structure, therefore adversely changing the physicochemical properties of LNPs. Consequently, a stability-indicating method capable of separating and detecting those low-level lipid degradants is required.
Separation is typically achieved by reverse-phase chromatography. Given the complex nature of functional excipient mixtures, UHPLC is preferred over HPLC due to its higher separation efficiency. Moreover, sharper peaks are highly desired for PEG-lipid in peak integration and accurate measurement considering its polydisperse nature.31
Lipids used for LNP possess low or no UV absorbance, thus an alternative detector such as a mass spectrometer or aerosol-based detection should be sought. Among those, the charged aerosol detector (CAD) has been widely employed due to its robustness, ease of use and availability.32,33 Figure 3 is an example of a stability indicating method used to separate and quantitate all lipid components and their major degradants and process-related impurities.
Conclusion
Analysis of OGNs, functional excipients and nanoparticle distribution/heterogeneity is critical in formulation development, product characterisation and stability monitoring for lipid nanoparticlebased OGNs. Work presented here highlights the application of liquid chromatography including AEX, IP-RPLC, SEC and UPLC-CAD in this area. Future advances will be heavily driven by the improvement of chromatography instrumentation, including both separation and detection, as well as the implementation of novel column technologies.
About the authors


Yong Liu is a Senior Principal Scientist in the Analytical Sciences department of MSD. Currently, he is leading a group in supporting drug product development. He completed his PhD at Rutgers University in organic chemistry. He has published 40 articles, with research interests in separation sciences, structural elucidation of drug molecule degradation impurities by mass spectrometry and the fundamental understanding of drug–drug and drug– excipient interactions.


Yuejie Zhao received her doctorate degree in Analytical Chemistry from the University of Georgia. She is currently a Senior Scientist in the Analytical Sciences department of MSD where she supports sterile formulation development focusing on novel therapeutic oligonucleotides delivery.


Adam Socia is an Associate Principal Scientist in the Vaccine Analytical Research and Development group at MSD. He completed his PhD at Drexel University, focusing on the development of novel separations for amino acids and other charged analytes. Adam’s current role is in providing analytical leadership for developmental vaccines and novel adjuvants. Adam has published his research in several peerreviewed journals and presented both oral and poster presentations at numerous conferences.


Andreas Abend received his doctorate degree in natural sciences from the University of Karlsruhe in Germany. He was a postdoc at the University of Wisconsin’s Enzyme Institute before starting his career with MSD. Andreas is currently a Director in the Analytical Sciences department of MSD leading a group of scientists supporting new drug product development.
References
- Capaldi D, Teasdale A, Henry S, Akhtar N, den Besten C, Gao-Sheridan S, Kretschmer M, Sharpe N, Andrews B, Burm B, Foy J. Impurities in Oligonucleotide Drug Substances and Drug Products, Nucleic Acid Ther. 27(6) (2017) 309-322.
- Chen D. Analytical chemistry in antisense oligonucleotide drug development, Front. Biotechnol. Pharm. 2 (2001) 202-224.
- Appling FD, Lucius AL, Schneider DA. Quantifying the influence of 5′-RNA modifications on RNA polymerase I activity. Biophys. Chem. 230 (2017) 84-88.
- Caton-Williams J, Lin L, Smith M, Huang Z. Convenient synthesis of nucleoside 5′-triphosphates for RNA transcription. Chemical Communications 47(28) (2011) 8142-8144.
- Lin L, Liu Q, Berube N, Detmer S, Zhou Y. 5′-triphosphate-short interfering RNA: potent inhibition of influenza A virus infection by gene silencing and RIG-I activation. J. Virol. 86(19) (2012) 10359-10369.
- Maitra U, Hurwitz H. The role of DNA in RNA synthesis, IX. Nucleoside triphosphate termini in RNA polymerase products. Proc Natl Acad Sci USA 54(3) (1965) 815-822.
- Seliger H, Frohlich A, Groger G, Krist B, Montenarh M, Rosch H, Rosch R, Ortigao FR. Synthetic oligonucleotides for biomedical applications. Nucleic Acids Symp Ser (24) (1991) 193-6.
- Vaghefi MM. Chemical synthesis of oligonucleotide triphosphates. CRC Press LLC, 2005, pp. 105-113.
- Zlatev I, Lackey JG, Zhang L, Dell A, McRae K, Shaikh S, Duncan RG, Rajeev KG, Manoharan M. Automated parallel synthesis of 5′-triphosphate oligonucleotides and preparation of chemically modified 5′-triphosphate small interfering RNA. Bioorg. Med. Chem. 21(3) (2013) 722-732.
- Zlatev I, Manoharan M, Vasseur J-J, Morvan F. Solidphase chemical synthesis of 5′-triphosphate DNA, RNA, and chemically modified oligonucleotides. Curr Protoc Nucleic Acid Chem Chapter 1 (2012) Unit1.28.
- Krotz AH, McElroy B, Scozzari AN, Cole DL, Ravikumar VT. Controlled Detritylation of Antisense Oligonucleotides. Org. Process Res. Dev. 7(1) (2003) 47-52.
- Srivatsa S, Klopchin P, Batt M, Feldman M, Carlson RH, Cole DL. Selectivity of anion exchange chromatography and capillary gel electrophoresis for the analysis of phosphorothioate oligonucleotides. J. Pharm. Biomed. Anal. 16(4) (1997) 619-630.
- Thayer J, Murugaiah V, Wu Y. Purity analysis and impurities determination by AEX-HPLC, CRC Press, 2011, pp. 47-104.
- Cramer H, Finn KJ, Herzberg E. Purity analysis and impurities determination by reversed-phase high-performance liquid chromatography, CRC Press, 2011, pp. 1-46.
- Levin DS, Shepperd BT, Gruenloh CJ. Combining ion pairing agents for enhanced analysis of oligonucleotide therapeutics by reversed phase-ion pairing ultra performance liquid chromatography (UPLC), J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 879(19) (2011) 1587-1595.
- Pourshahian S, McCarthy SM. Analysis of oligonucleotides by liquid chromatography and mass spectrometry, CRC Press, 2011, pp. 137-166.
- Roussis SG, Cedillo I, Rentel C. Semi-quantitative determination of co-eluting impurities in oligonucleotide drugs using ion-pair reversed-phase liquid chromatography mass spectrometry, J. Chromatogr. A 1584 (2019) 106-114.
- Roussis SG, Pearce M, Rentel C. Small alkyl amines as ion-pair reagents for the separation of positional isomers of impurities in phosphate diester oligonucleotides, J. Chromatogr. A 1594 (2019) 105-111.
- Li S, Lu D, Zhang Y, Wang S. An Improved Ion-Pair Reversed Phase LC Method for Analysis of Major Impurities of Phosphorothioate Oligonucleotide Cantide, Chromatographia 72 (2010) 215-223.
- Pan S, Shi Y. Recent advances in the characterization and analysis of therapeutic oligonucleotides by analytical separation methods coupling with mass spectrometry. Adv. Chromatogr. (Boca Raton, FL, U. S.) 53 (2017) 143-177.
- Gregoriadis G. Liposome technology (Vol. 3), CRC press 1992.
- Kasai K. Size-dependent chromatographic separation of nucleic acids, Journal of Chromatography B: Biomedical Sciences and Applications 618(1-2) (1993) 203-221.
- Podzimek S. Light scattering, size exclusion chromatography and asymmetric flow field flow fractionation: powerful tools for the characterization of polymers, proteins and nanoparticles, John Wiley & Sons 2011.
- Zhang J, Haas RM, Leone AM. Polydispersity characterization of lipid nanoparticles for siRNA delivery using multiple detection size-exclusion chromatography, 84(14) (2012) 6088-6096.
- Zhang J, Pei Y, Zhang H, Wang L, Arrington L, Zhang Y, Glass A, Leone AM. Assessing the heterogeneity level in lipid nanoparticles for siRNA delivery: size-based separation, compositional heterogeneity, and impact on bioperformance, 10(1) (2013) 397-405.
- Liu C, Liu G, Liu N, Zhang Y, He J, Rusckowski M, Hnatowich DJ. Radiolabeling morpholinos with 90Y, 111In, 188Re and 99mTc, Nucl. Med. Biol. 30(2) (2003) 207-214.
- Li W, Szoka FC. Lipid-based nanoparticles for nucleic acid delivery, 24(3) (2007) 438-449.
- Stanton MG, Colletti SL. Medicinal chemistry of siRNA delivery, 53(22) (2010) 7887-7901.
- Beckwith ALJ, Eichinger PH, Mooney BA, Prager RH. Amine autoxidation in aqueous solution. Australian Journal of Chemistry 36(4) (1983) 719-739.
- Grit M, de Smidt JH, Struijke A, Crommelin DJA. Hydrolysis of phosphatidylcholine in aqueous liposome dispersions. International Journal of Pharmaceutics 50(1) (1989) 1-6.
- Li Z, Schariter JA, Zhang J, Davis JC, Leone AM. Application of ultra-high performance liquid chromatography for chemical characterization of liposome-based therapeutic small-interfering RNA. Am. Pharm. Rev. 13(6) (2010) 102, 104, 106-108, 110.
- Almeling S, Ilko D, Holzgrabe U. Charged aerosol detection in pharmaceutical analysis, 69 (2012) 50-63.
- Cohen RD, Liu Y. Advances in aerosol-based detectors. Adv. Chromatogr. (Boca Raton, FL, U. S.) 52 (2014).
Issue
Related topics
Analytical techniques, Chromatography, Drug Delivery Systems, Excipients, QA/QC










In RPLC analysis i have one question. It is very nice work, and simplified the analysis by quantifying using LC methods. I wondered how the nucleic acids will be protected without the addition of protecting buffers in sample preparations. Is any way to share the spectral purity in each analysis? Did you obtained any stability of the samples.